Imperial/Infector Detector/Testing
From 2007.igem.org

Infector Detector: Testing
Summary
The key results of the testing were:
- The optimum DNA concentration for [http://partsregistry.org/Part:BBa_T9002 pTet-LuxR-pLux-GFPmut3b] in our Commcercial S30 Cell extract is 4µg.
Aims
The aims of the testing were as follows:
- To test and obtain the optimal DNA concentration for construct 1 in vitro
- To characterise the output of GFPmut3b for a range of AHL inputs. From this obtain the AHL sensitivity of our system.
In addition the fluorescence measurements were converted to number of GFPmut3b molecules synthesised using a calibration curve constructed using purified GFPmut3b.
Results
DNA Concentrations
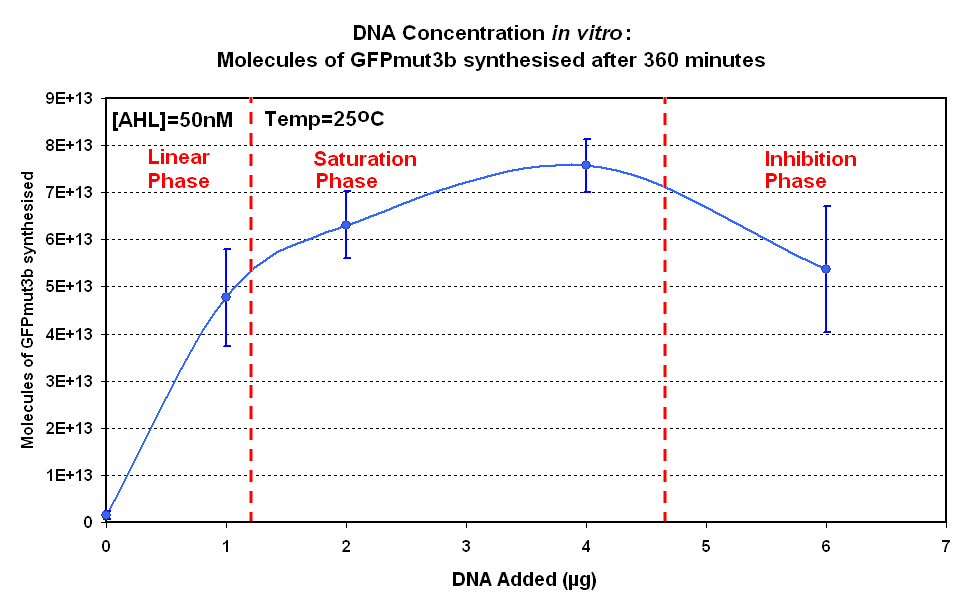
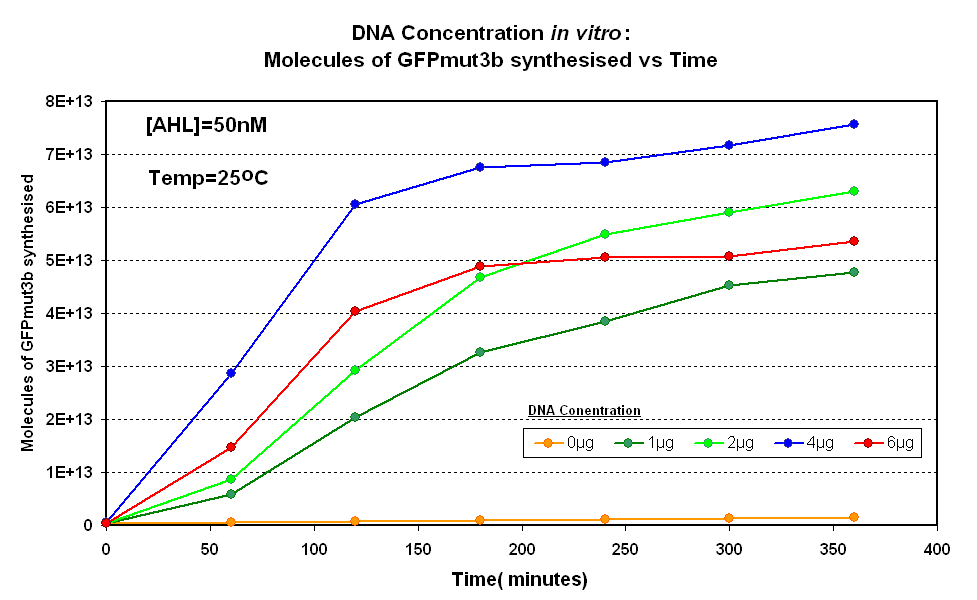 Fig.1.1:Molecules of GFPmut3b synthesised over time, for each DNA Concentration in vitro - The fluorescence was measured over time for each experiment and converted into molecules of GFPmut3b in vitro using our calibration curve Click here for results and protocol. |
The Results above show that the optimum DNA concentration for in vitro is 4µg for 50nM AHL. From figure 1.1. and 1.2 it can be seen that as DNA concentration increases above 4µg the GFPmut3b molecules synthesised decrease. Interestingly for figure 1.2 the graph can be split into several regions of how the DNA concentration changes the output of GFPmut3b synthesis:
- Linear phase - The initial relationship between GFP output and DNA concentration is linear
- Saturation phase - The affect of the DNA concentration on GFP synthesis levels off and peaks at 4µg.
- Inhibition phase- Increasing the DNA concentration actually inhibits the rate of protein synthesis.
The fact that increasing DNA concentration above 4µg causes a decrease in rate of protein synthesis is very interesting. The reason for this is thought to be that increasing DNA concentration causes problems with premature translational termination.
For more information on the results please go the the results page
The 4µg was taken as the maximum and used for the rest of the testing.
AHL Testing
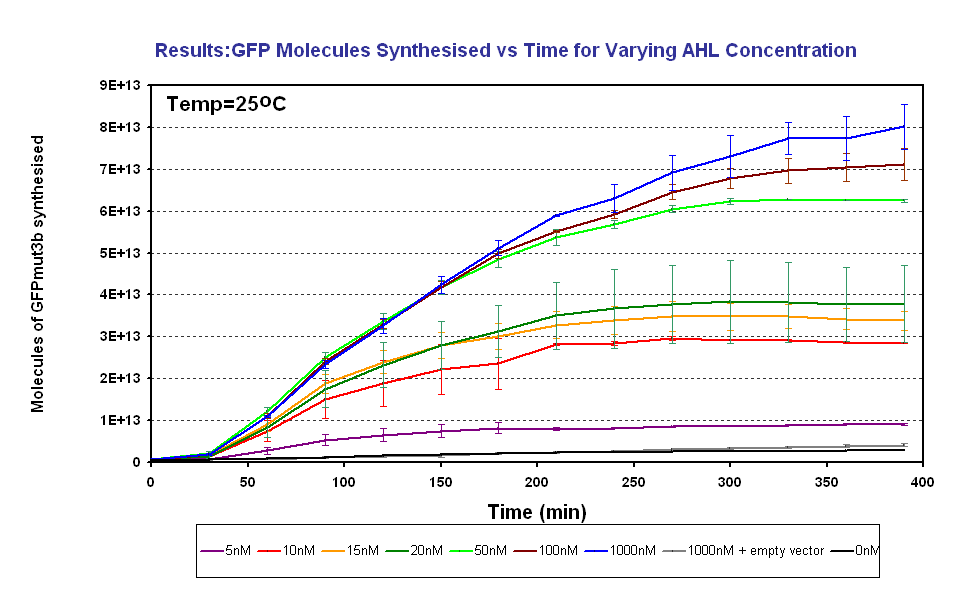
Figure 1.3 shows us the following:
- The output of GFPmut3b increases with input of AHL
- The system is sensitive to a range of 5-1000nM AHL
- The GFPmut3b molecules synthesis stops at ~300minutes. This could be due to steady state or due to no synthesis of GFPmut3b. It is known not to be steady state because the degradation experiment(link) proved degradation is negligible. Interestingly this time is independent of the GFPmut3b molecules produced, showing that the LuxR under the control of pTet is the major source of energy consumption. This highlights the advantages of using the construct 2 [http://partsregistry.org/Part:BBa_J37032 pLux-GFPmut3b] that does not have the energetic burden of producing LuxR
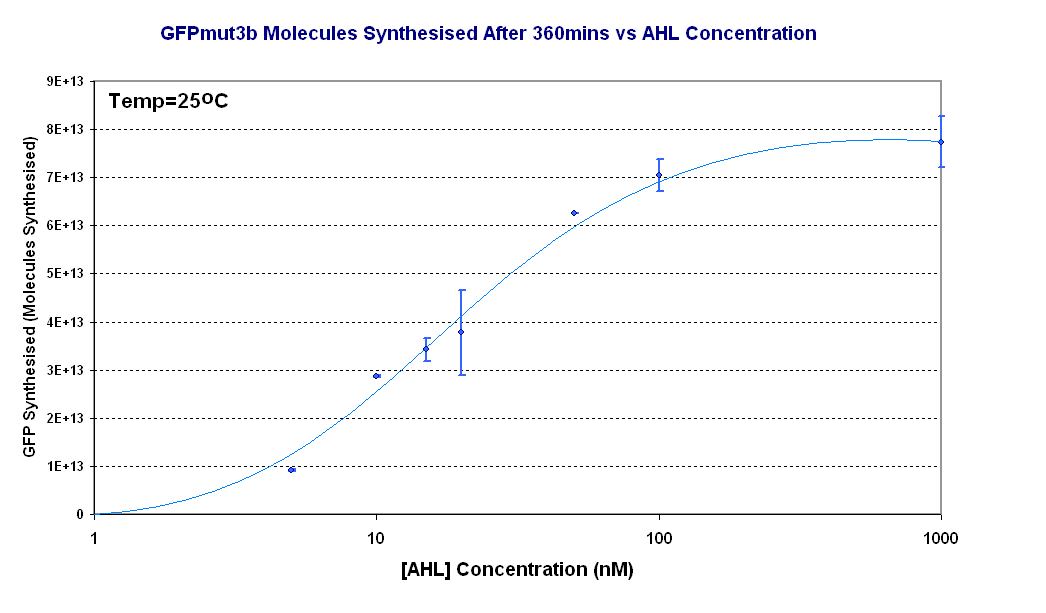
Figure 1.4 shows the Transfer Function of the construct 1 in vitro. From this graph the following information can be extracted: