E-Notebook
From 2007.igem.org
(→This Week) |
|||
| (34 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {|align="justify" | |
| - | [[Image: | + | |On June 1st 2007, a group of undergraduates of various disciplines and from around the country assembled at NCBS, to attempt a 'proof of principle' demonstration. Their mission: to assemble and test a 'genetically engineered machine', a complex network assembled from simple biological parts. You will be amazed how much they managed to achieve in just six weeks! |
| - | + | |[[Image:Ncbs_Logo.jpg|200px|center]] | |
|- | |- | ||
| - | | | + | | |
| - | | | + | ''The National Centre for Biological Sciences is a research institute located in Bangalore, India. In 2006, a team of graduate students from NCBS was the first from India to participate in iGEM. This year, we are very excited to compete as an all-undergraduate team of six students, from six different colleges located in four cities around India. Now that the summer is over, we hope to take the excitement of iGEM back to our home institutions, and create new teams across India who will participate in future iGEMs.'' |
| - | + | |[[Image:Ncbs.jpg|400px|center]] | |
| - | + | |- | |
| - | + | | | |
| - | + | |align="center"|[http://www.ncbs.res.in/ National Centre for Biological Sciences, Bangalore] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | + | <!--- The Mission, Experiments ---> | |
| - | + | ||
| - | + | ||
| + | {| style="color:#294e9c;background-color:#e1e0db;" cellpadding="3" cellspacing="1" border="1" bordercolor="#1100ff" width="62%" align="center" | ||
| + | !align="center"|[[Bangalore|Home]] | ||
| + | !align="center"|[[The Team|The Team]] | ||
| + | !align="center"|[[The Goal|The Goal]] | ||
| + | !align="center"|[[Parts Data Sheets|Parts Data Sheets]] | ||
| + | !align="center"|[[Mathematical Models|Mathematical Models]] | ||
| + | !align="center"|[[e-Notebook|e-Notebook]] | ||
| + | |} | ||
| - | + | ==e-Notebook == | |
| - | + | {| align="center" border="0" cellpadding="5" cellspacing="3" | |
| - | + | !colspan="7"|June 07 | |
| - | == | + | !colspan="7"| |
| - | {| align="center" border="0" cellpadding="5" cellspacing="3" | + | !colspan="7"|July 07 |
| - | + | ||
| - | + | ||
| - | !colspan="7"|June 07 | + | |
| - | !colspan="7"| | + | |
| - | !colspan="7"|July 07 | + | |
|- | |- | ||
| - | |'''S''' | + | |'''S''' |
| - | |M | + | |M |
| - | |T | + | |T |
| - | |W | + | |W |
| - | |T | + | |T |
| - | |F | + | |F |
| - | |S | + | |S |
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |'''S''' | ||
| + | |M | ||
| + | |T | ||
| + | |W | ||
| + | |T | ||
| + | |F | ||
| + | |S | ||
| + | |- | ||
| | | | ||
| | | | ||
| Line 46: | Line 58: | ||
| | | | ||
| | | | ||
| - | | | + | |[[June 1|1]] |
| - | | | + | |2 |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | | | + | |[[July 1|1]] |
| - | | | + | |[[July 2|2]] |
| - | | | + | |[[July 3|3]] |
| - | | | + | |[[July 4|4]] |
| - | | | + | |[[July 5|5]] |
| - | | | + | |[[July 6|6]] |
| - | | | + | |[[July 7|7]] |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | + | |3 | |
| - | + | |[[June 4|4]] | |
| - | + | |[[June 5|5]] | |
| - | + | |[[June 6|6]] | |
| - | |3 | + | |[[June 7|7]] |
| - | |4 | + | |[[June 8|8]] |
| - | |5 | + | |9 |
| - | | | + | | |
| - | | | + | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | |[[July 8|8]] | |
| - | + | |[[July 9|9]] | |
| - | + | |[[July 10|10]] | |
| - | + | |[[July 11|11]] | |
| - | | | + | |[[July 12|12]] |
| - | |[[June | + | |[[July 13|13]] |
| - | |[[June | + | |[[July 14|14]] |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | |[[July | + | |
| - | |[[July | + | |
| - | |[[July | + | |
| - | |[[July | + | |
| - | |[[July | + | |
| - | |[[July | + | |
| - | |[[July | + | |
|- | |- | ||
| - | + | |10 | |
| - | + | |[[June 11|11]] | |
| - | + | |[[June 12|12]] | |
| - | + | |[[June 13|13]] | |
| - | + | |[[June 14|14]] | |
| - | + | |[[June 15|15]] | |
| - | + | |[[June 16|16]] | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | |[[July 15|15]] | |
| - | + | |16 | |
| - | + | |17 | |
| - | + | |18 | |
| - | + | |19 | |
| - | + | |20 | |
| - | + | |21 | |
| - | + | |- | |
| - | + | |[[June 17|17]] | |
| - | + | |[[June 18|18]] | |
| - | + | |[[June 19|19]] | |
| - | + | |[[June 20|20]] | |
| - | + | |[[June 21|21]] | |
| - | + | |[[June 22|22]] | |
| - | + | |[[June 23|23]] | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | |22 | |
| - | + | |23 | |
| - | + | |24 | |
| - | + | |25 | |
| - | + | |26 | |
| - | + | |27 | |
| - | + | |28 | |
| - | + | |- | |
| - | + | |[[June 24|24]] | |
| - | + | |[[June 25|25]] | |
| - | + | |[[June 26|26]] | |
| - | + | |[[June 27|27]] | |
| - | + | |[[June 28|28]] | |
| - | + | |[[June 29|29]] | |
| - | |10 | + | |[[June 30|30]] |
| - | |[[June 11|11]] | + | | |
| - | |[[June 12|12]] | + | | |
| - | |[[June 13|13]] | + | | |
| - | |[[June 14|14]] | + | | |
| - | |[[June 15|15]] | + | | |
| - | |[[June 16|16]] | + | | |
| - | | | + | | |
| - | | | + | |29 |
| - | | | + | |30 |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | | | + | | |
| - | |[[July 15|15]] | + | | |
| - | |16 | + | |
| - | |17 | + | |
| - | |18 | + | |
| - | |19 | + | |
| - | |20 | + | |
| - | |21 | + | |
| - | |- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |[[June 17|17]] | + | |
| - | |[[June 18|18]] | + | |
| - | |[[June 19|19]] | + | |
| - | |[[June 20|20]] | + | |
| - | |[[June 21|21]] | + | |
| - | |[[June 22|22]] | + | |
| - | |[[June 23|23]] | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | |22 | + | |
| - | |23 | + | |
| - | |24 | + | |
| - | |25 | + | |
| - | |26 | + | |
| - | |27 | + | |
| - | |28 | + | |
| - | |- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |[[June 24|24]] | + | |
| - | |[[June 25|25]] | + | |
| - | |[[June 26|26]] | + | |
| - | |[[June 27|27]] | + | |
| - | |[[June 28|28]] | + | |
| - | |[[June 29|29]] | + | |
| - | |[[June 30|30]] | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | |29 | + | |
| - | |30 | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|} | |} | ||
| + | The following is the record of all the experiments done by us, each followed by graphs obtained by analysis of the corresponding microscopy and flow cytometry data. Note that, with flow cytometry, signals obtained from each filter do not exactly correspond to CFP or YFP amount inside a cell, because of spectral overlap of their excitation and emission spectra. We used a [[Media:Analysis.pdf|linear transformation]] to separate CFP and YFP from autofluorescence and noise. | ||
| + | |||
| + | == Experiment == | ||
| + | |||
| + | === Equivalences === | ||
| + | |||
| + | *pL.Cfp | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:pLC_dup.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pT.luxI.Cfp | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:pTIC_dup.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pL.luxI.Cfp | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:pLIC_dup.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pL.luxR.Yfp | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:pLRY_dup.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | === Open loops === | ||
| - | = | + | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (0 ng/ml aTc) |
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:Openloop_0.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (1 ng/ml aTc) | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:Openloop_1.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (5 ng/ml aTc) | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:Openloop_5.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (10 ng/ml aTc) | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:Openloop_10.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (20 ng/ml aTc) | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:Openloop_20.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (50 ng/ml aTc) | ||
| + | {| | ||
| + | |- align="justify" | ||
| + | |[[Image:Openloop_50.png|400px]] | ||
| + | |Details... | ||
| + | |} | ||
| + | |||
| + | === Closed loops === | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =='''Protocols''' == | ||
'''1) Induction experiments:''' | '''1) Induction experiments:''' | ||
| Line 264: | Line 257: | ||
'''2) Open loop experiments:''' | '''2) Open loop experiments:''' | ||
| - | i) Morning imaging | + | i) Morning imaging (followed usually) |
Day 0: 0000: Inoculate pTLuxI.C in LB | Day 0: 0000: Inoculate pTLuxI.C in LB | ||
| - | Day 1: | + | Day 1: 0100: Transfer to Glu M9 in 250 mL flask with a particular [aTc] and inoculum [x3] |
1230: Inoculate pLLuxR.Y pRC in LB | 1230: Inoculate pLLuxR.Y pRC in LB | ||
2200: Filter required OD flask | 2200: Filter required OD flask | ||
2230: Make 7 dilutions for pLLuxR.YpRC with 50 old:50 new 2xM9 | 2230: Make 7 dilutions for pLLuxR.YpRC with 50 old:50 new 2xM9 | ||
-ve control: K12z1 | -ve control: K12z1 | ||
| - | Day 2: | + | Day 2: 1000: Take microscopy images |
ii) Night imaging | ii) Night imaging | ||
Day 1: 1100: Inoculate pTLuxI.C in LB | Day 1: 1100: Inoculate pTLuxI.C in LB | ||
| Line 281: | Line 274: | ||
2130: Take microscopy images | 2130: Take microscopy images | ||
| + | ==Notes== | ||
| + | *All inductions with IPTG: 0,5,10,50,100,500,1000 uM | ||
| + | |||
| + | *All inductions with aTc: 0,1,5,10,20,50 ng/mL | ||
| + | *The open loop experiments to be carried out with following aTc concentrations: | ||
| - | == Experiments | + | [A] : 20 ng/ml |
| + | |||
| + | [B] : 10 ng/ml | ||
| + | |||
| + | [C] : 5 ng/ml | ||
| + | |||
| + | [D] : 1 ng/ml | ||
| + | |||
| + | [E] : 50 ng/ml | ||
| + | |||
| + | [F] : 0 ng/ml | ||
| + | |||
| + | [AA] : 20 ng/ml (repeat A) | ||
| + | |||
| + | [FF] : 0 ng/ml (repeat F) | ||
| + | |||
| + | |||
| + | The '''''DUPLICATES''''' for each of the above experiments are denoted by [Aa], [Bb], [Cc], [Dd], [Ee] and [Ff] respectively. | ||
| + | |||
| + | == This Week == | ||
| + | |||
| + | {|- | ||
| + | | | ||
| + | Mon, July 2: pL LuxI.C for IPTG-aTc equivalence (morn) | ||
| + | Tue, July 3: pL LuxR.Y for cfp-yfp equivalence, | ||
| + | pT LuxI.C for IPTG-aTc | ||
| + | Wed, July 4: Open loop [E],[F] (aTc) | ||
| + | At OD 0.1: To prove scaling argument for AI. | ||
| + | Thu, July 5: aTC 10,20 ng/mL [X,Y] | ||
| + | DUPLICATES: | ||
| + | --------- | ||
| + | At OD 0.2: | ||
| + | Fri, July 6: Open loop: aTC 20,10 ng/mL [A',B'] | ||
| + | Sat, July 7: Open loop: aTC 5,1 ng/mL [C',D'] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | == Experiments Record == | ||
*Calibration of the number of colonies obtained at different optical densities. | *Calibration of the number of colonies obtained at different optical densities. | ||
*Transformation of competent E.coli (strain K12Z1) cells with constructs. | *Transformation of competent E.coli (strain K12Z1) cells with constructs. | ||
| Line 457: | Line 493: | ||
*'''The Scaling Argument''' | *'''The Scaling Argument''' | ||
| - | Two sets of experiments would be carried out, where cells would be grown to an optical density of | + | Two sets of experiments would be carried out, where cells would be grown to an optical density of 0.257 & 0.015 respectively in order to check for density dependence of autoinducer production. The supernatent would be filtered out and the one from the OD=0.257 would be scaled down to OD=0.015. The cells would then be imaged using a phase contrast microscope. The scaling argument would be proved if we get the same cfp expression from both. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Discussions == | == Discussions == | ||
Latest revision as of 07:22, 24 October 2007
| On June 1st 2007, a group of undergraduates of various disciplines and from around the country assembled at NCBS, to attempt a 'proof of principle' demonstration. Their mission: to assemble and test a 'genetically engineered machine', a complex network assembled from simple biological parts. You will be amazed how much they managed to achieve in just six weeks! | |
|
The National Centre for Biological Sciences is a research institute located in Bangalore, India. In 2006, a team of graduate students from NCBS was the first from India to participate in iGEM. This year, we are very excited to compete as an all-undergraduate team of six students, from six different colleges located in four cities around India. Now that the summer is over, we hope to take the excitement of iGEM back to our home institutions, and create new teams across India who will participate in future iGEMs. | |
| [http://www.ncbs.res.in/ National Centre for Biological Sciences, Bangalore] |
| Home | The Team | The Goal | Parts Data Sheets | Mathematical Models | e-Notebook |
|---|
Contents |
e-Notebook
| June 07 | July 07 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | M | T | W | T | F | S | S | M | T | W | T | F | S | |||||||
| 1 | 2 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||||||||||
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |||||||
| 10 | 11 | 12 | 13 | 14 | 15 | 16 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |||||||
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |||||||
| 24 | 25 | 26 | 27 | 28 | 29 | 30 | 29 | 30 | ||||||||||||
The following is the record of all the experiments done by us, each followed by graphs obtained by analysis of the corresponding microscopy and flow cytometry data. Note that, with flow cytometry, signals obtained from each filter do not exactly correspond to CFP or YFP amount inside a cell, because of spectral overlap of their excitation and emission spectra. We used a linear transformation to separate CFP and YFP from autofluorescence and noise.
Experiment
Equivalences
- pL.Cfp
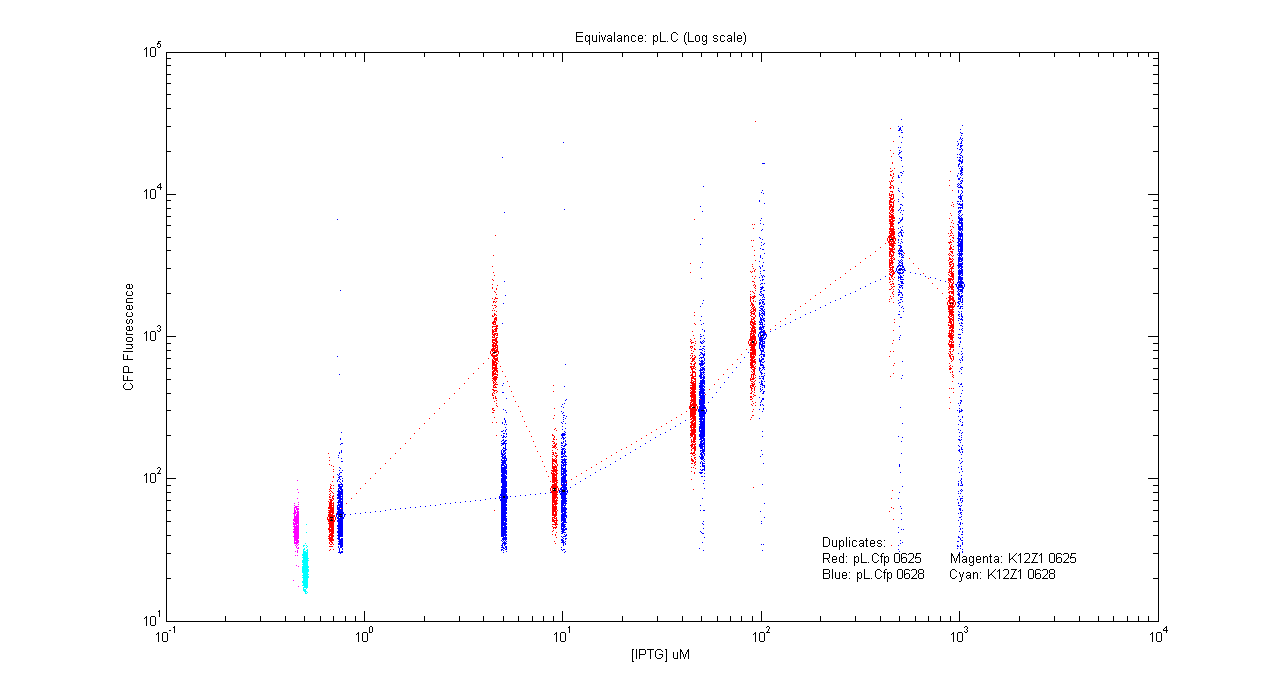
| Details... |
- pT.luxI.Cfp
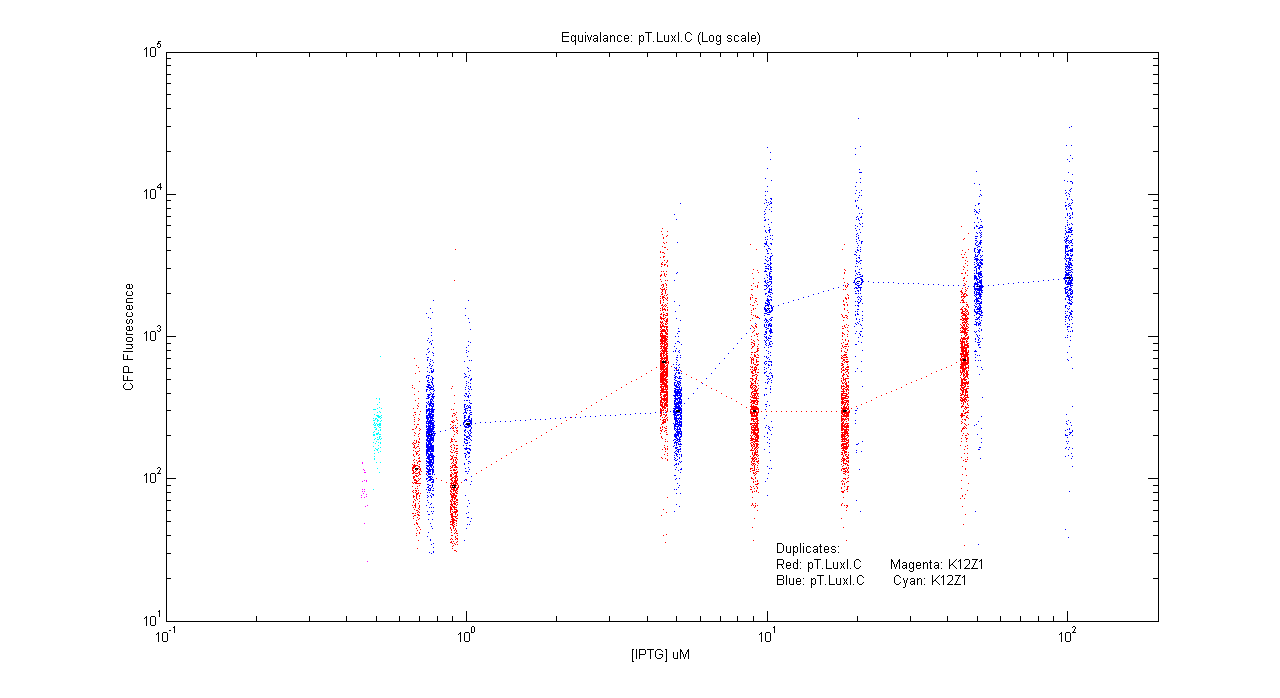
| Details... |
- pL.luxI.Cfp
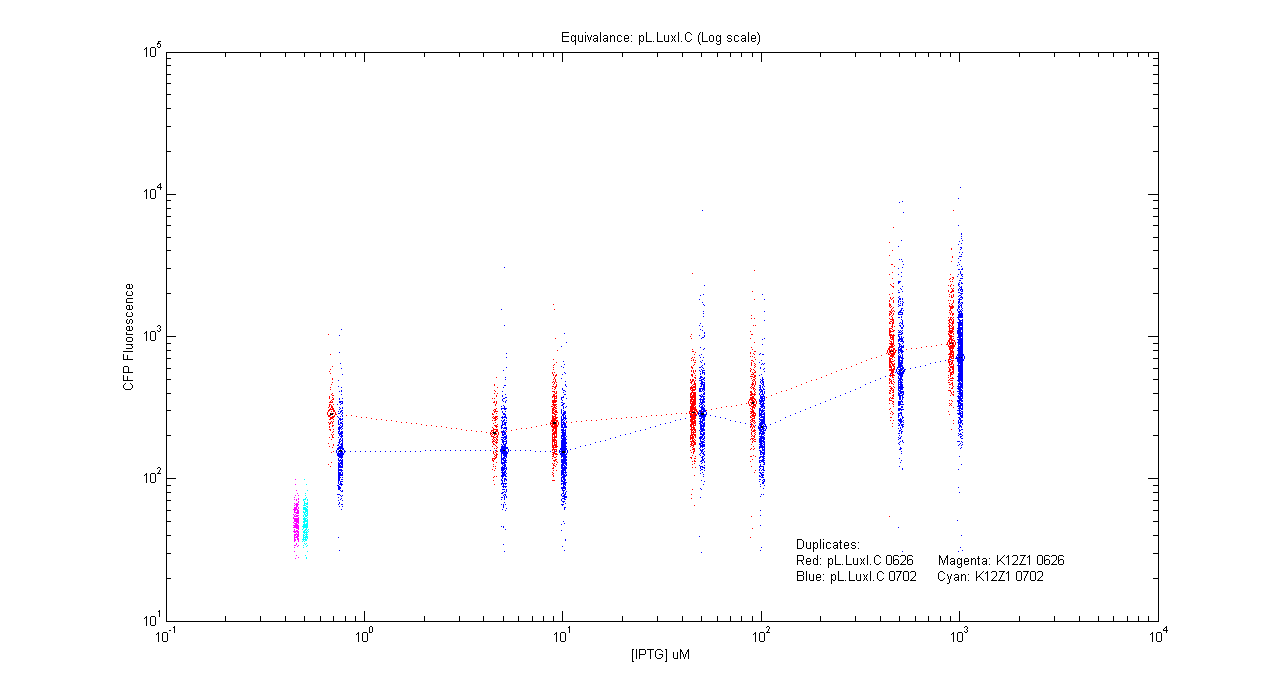
| Details... |
- pL.luxR.Yfp
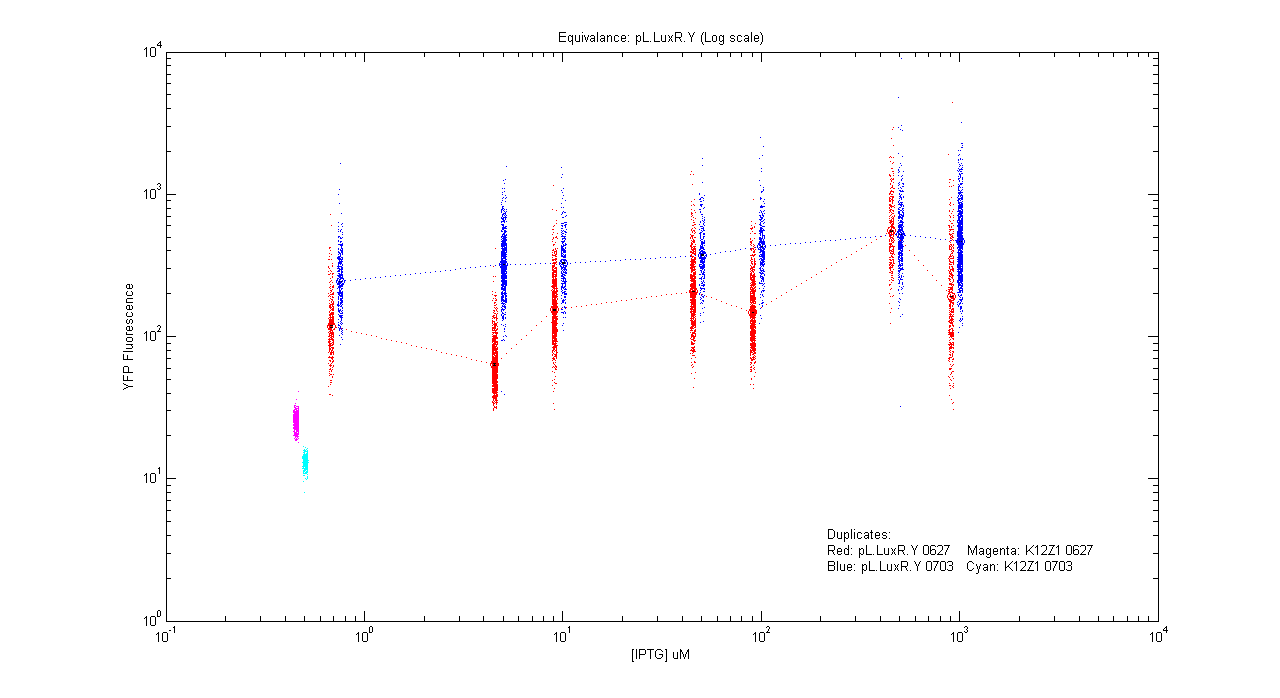
| Details... |
Open loops
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (0 ng/ml aTc)
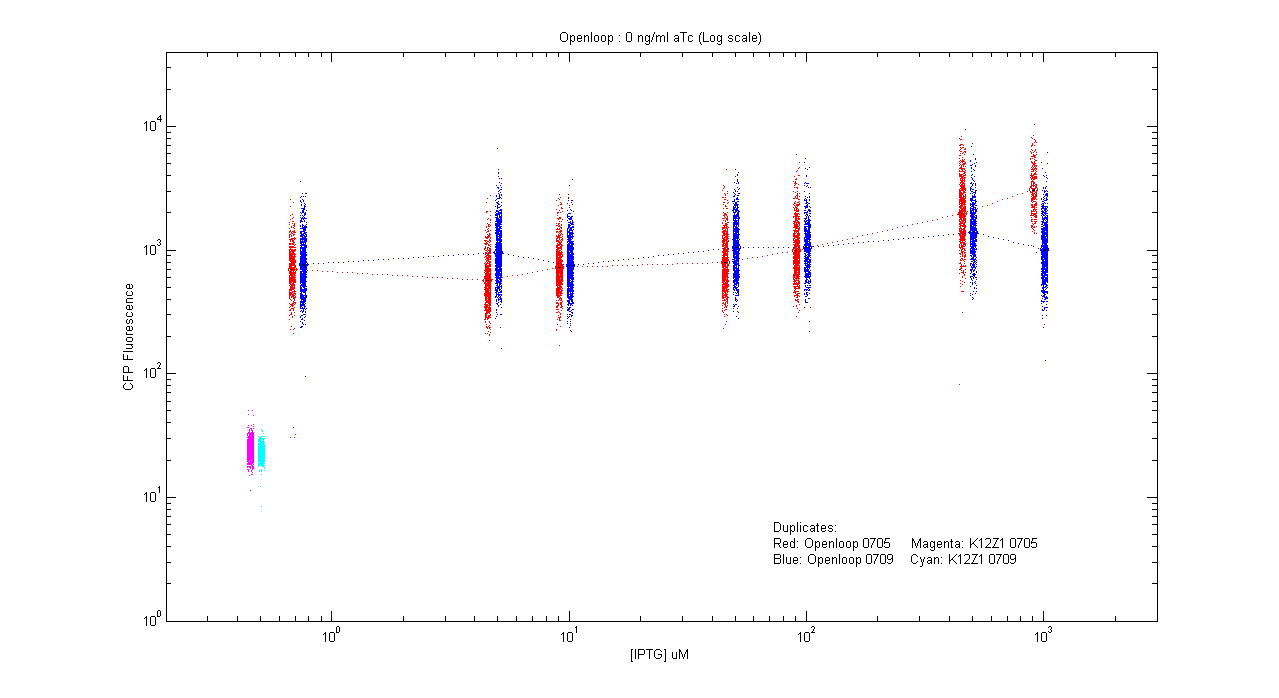
| Details... |
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (1 ng/ml aTc)
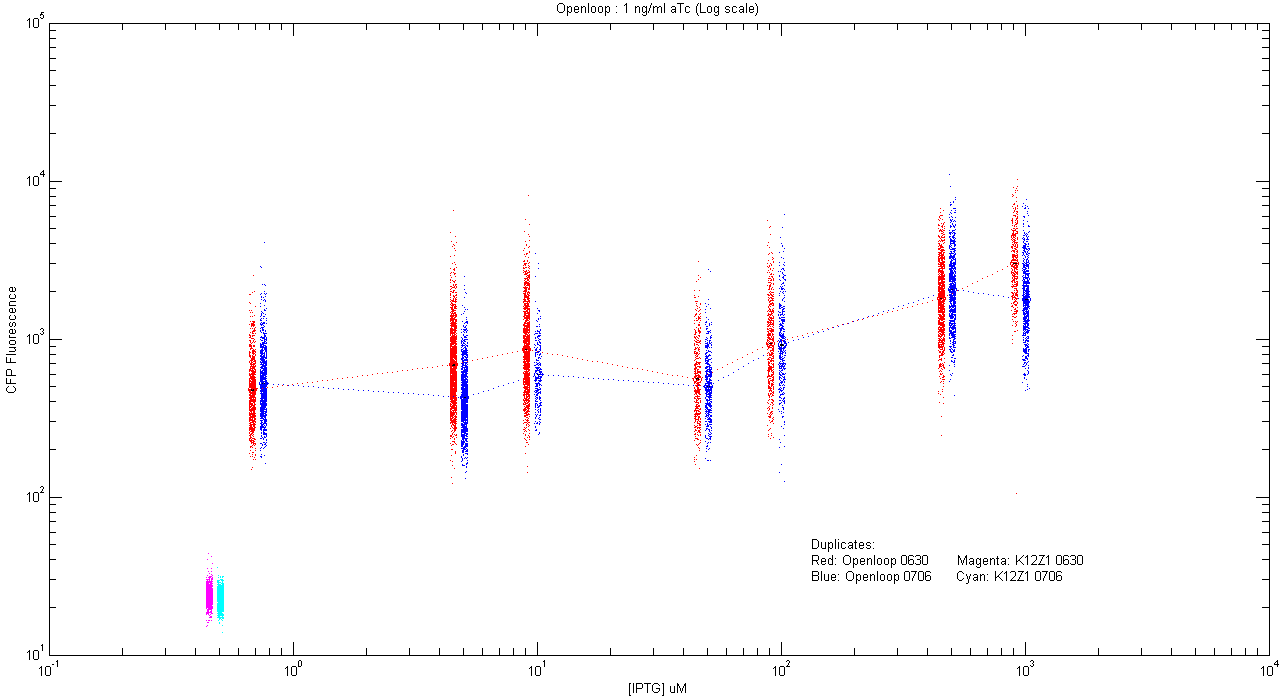
| Details... |
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (5 ng/ml aTc)
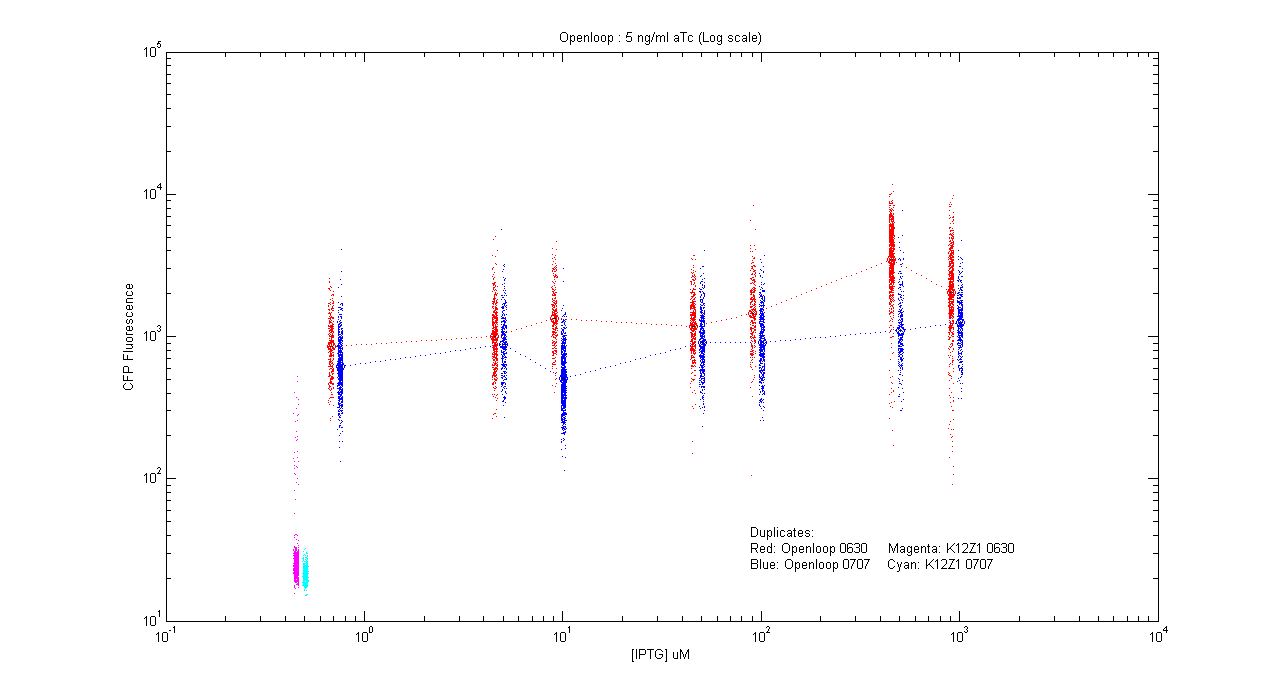
| Details... |
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (10 ng/ml aTc)
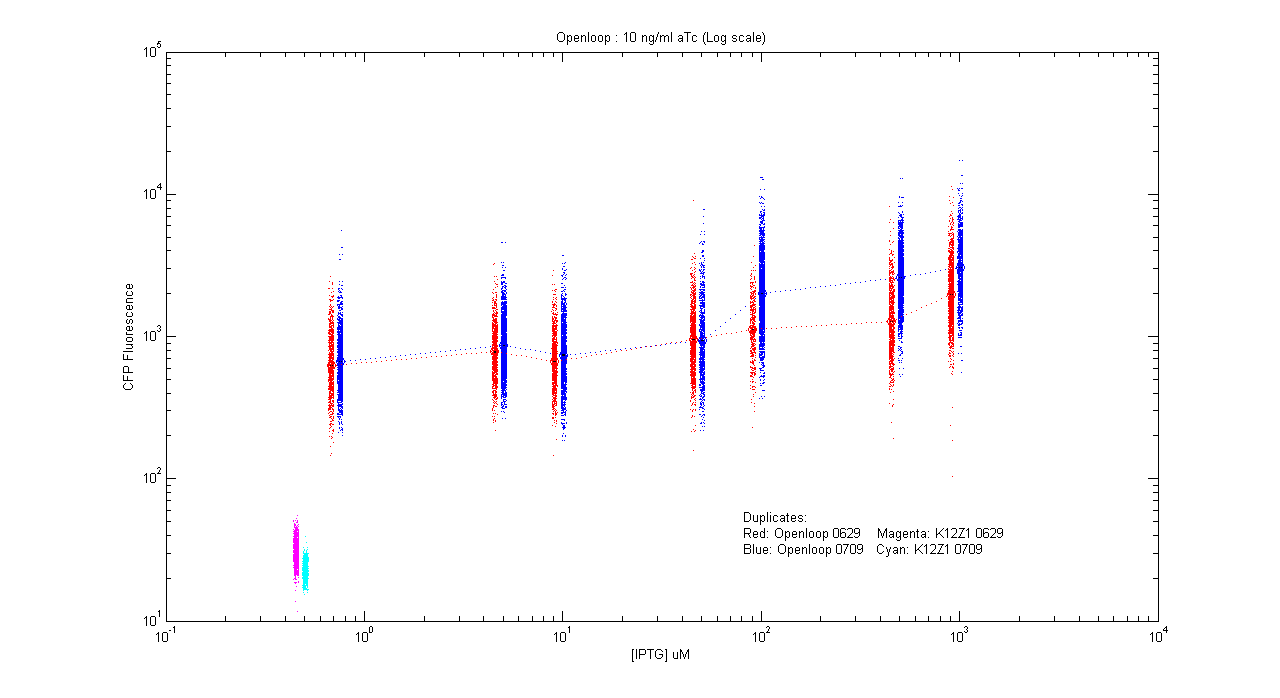
| Details... |
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (20 ng/ml aTc)
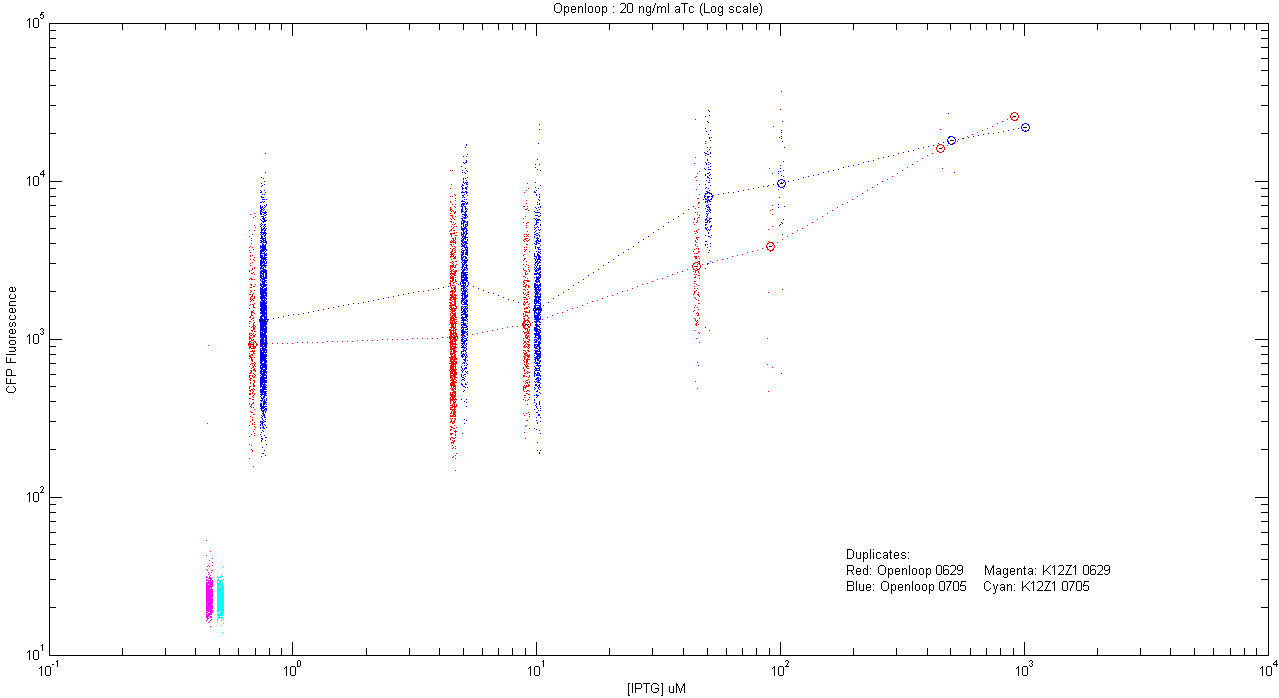
| Details... |
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (50 ng/ml aTc)
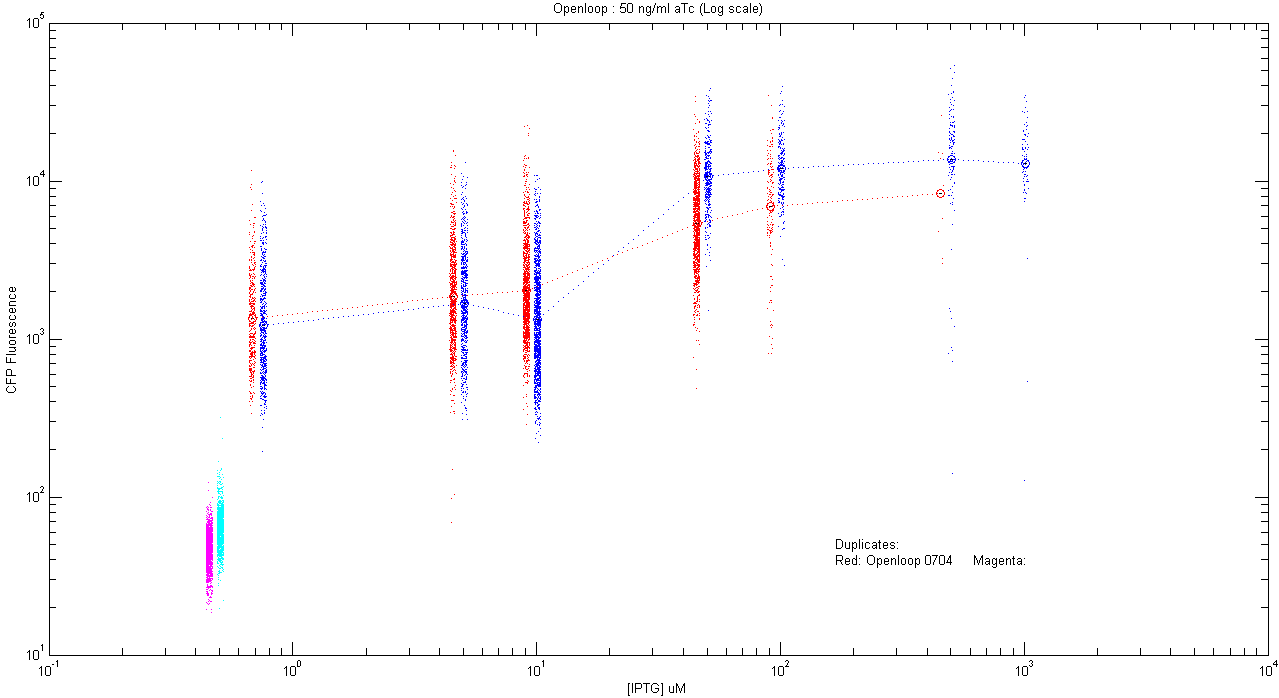
| Details... |
Closed loops
Protocols
1) Induction experiments:
Grow in LB for 10 hours
Then, induce in Glu M9 for 12 hours at various concns of IPTG/aTc
+ve control: pL YFP in 50,100 uM IPTG
-ve control: K12z1
Take microscopy images
2) Open loop experiments:
i) Morning imaging (followed usually)
Day 0: 0000: Inoculate pTLuxI.C in LB
Day 1: 0100: Transfer to Glu M9 in 250 mL flask with a particular [aTc] and inoculum [x3]
1230: Inoculate pLLuxR.Y pRC in LB
2200: Filter required OD flask
2230: Make 7 dilutions for pLLuxR.YpRC with 50 old:50 new 2xM9
-ve control: K12z1
Day 2: 1000: Take microscopy images
ii) Night imaging
Day 1: 1100: Inoculate pTLuxI.C in LB
2100: Transfer to Glu M9 in 250 mL flask with a particular [aTc] and inoculum [x3]
2330: Inoculate pLLuxR.YpRC in LB
Day 2: 0900: Filter required OD flask
0930: Make 7 dilutions for pLLuxR.YpRC with 50 old:50 new 2xM9
-ve control: K12z1 in 50 old:50 new 2xM9
2130: Take microscopy images
Notes
- All inductions with IPTG: 0,5,10,50,100,500,1000 uM
- All inductions with aTc: 0,1,5,10,20,50 ng/mL
- The open loop experiments to be carried out with following aTc concentrations:
[A] : 20 ng/ml
[B] : 10 ng/ml
[C] : 5 ng/ml
[D] : 1 ng/ml
[E] : 50 ng/ml
[F] : 0 ng/ml
[AA] : 20 ng/ml (repeat A)
[FF] : 0 ng/ml (repeat F)
The DUPLICATES for each of the above experiments are denoted by [Aa], [Bb], [Cc], [Dd], [Ee] and [Ff] respectively.
This Week
Mon, July 2: pL LuxI.C for IPTG-aTc equivalence (morn)
Tue, July 3: pL LuxR.Y for cfp-yfp equivalence,
pT LuxI.C for IPTG-aTc
Wed, July 4: Open loop [E],[F] (aTc)
At OD 0.1: To prove scaling argument for AI.
Thu, July 5: aTC 10,20 ng/mL [X,Y]
DUPLICATES:
---------
At OD 0.2:
Fri, July 6: Open loop: aTC 20,10 ng/mL [A',B']
Sat, July 7: Open loop: aTC 5,1 ng/mL [C',D']
|
Experiments Record
- Calibration of the number of colonies obtained at different optical densities.
- Transformation of competent E.coli (strain K12Z1) cells with constructs.
- Induction of the construct pLac cfp
E.coli cells transformed with the construct pLac cfp were cultured for 10 hours in Luria Bertani medium containing 50mg/ml Ampicillin.
The cells were then transferred into a minimal medium (M9 medium) and were induced with the following concentrations of IPTG. A stock concentration of 100mM was used.
| Sample | 100mM IPTG (ul) | 1mM IPTG (ul) | Inoculum volume from LB (ul) | Volume of M9 added (ml) |
|---|---|---|---|---|
| pLac cfp (0 uM) | - | - | 3 ul | 3 ml |
| pLac cfp (1 uM) | - | 3 ul | 3 ul | 3 ml |
| pLac cfp (10 uM) | - | 30 ul | 3 ul | 3 ml |
| pLac cfp (50 uM) | 1.5 ul | - | 3 ul | 3 ml |
| pLac cfp (100 uM) | 3 ul | - | 3 ul | 3 ml |
| pLac cfp (1000 uM) | 30 ul | - | 3 ul | 3 ml |
| K12Z1 (Autoflourescence) | - | - | 3 ul | 3 ml |
Cells were allowed to grow till the optical density was in the range of 0.05-0.1 (early exponential phase) and were imaged using a phase contrast microscope.
Results: The mean fluorescence of CFP was seen to increase with increasing concentrations of the IPTG.
- Induction of the construct pLac yfp
E.coli cells transformed with the construct pLac yfp were cultured for 10 hours in Luria Bertani medium containing 50mg/ml Ampicillin.
The cells were then transferred into a minimal medium (M9 medium) and were induced with the following concentrations of IPTG. A stock concentration of 100mM was used.
| Sample | 100mM IPTG (ul) | 1mM IPTG (ul) | Inoculum volume from LB (ul) | Volume of M9 added (ml) |
|---|---|---|---|---|
| pLac yfp (0 uM) | - | - | 3 ul | 3 ml |
| pLac yfp (1 uM) | - | 3 ul | 3 ul | 3 ml |
| pLac yfp (10 uM) | - | 30 ul | 3 ul | 3 ml |
| pLac yfp (50 uM) | 1.5 ul | - | 3 ul | 3 ml |
| pLac yfp (100 uM) | 3 ul | - | 3 ul | 3 ml |
| pLac yfp (1000 uM) | 30 ul | - | 3 ul | 3 ml |
| K12Z1 (Autoflourescence) | - | - | 3 ul | 3 ml |
Results: The mean fluorescence of YFP was seen to increase with increasing concentrations of the IPTG.
- Induction of the construct pTet.luxI.cfp
E.coli cells transformed with the construct pTet.luxI.cfp were cultured for 10 hours in Luria Bertani medium containing 50mg/ml Ampicillin.
The cells were then transferred into a minimal medium (M9 medium) and were induced with the following concentrations of aTc. A stock concentration of 1mg/ml was used.
| Sample | 10ug/ml aTc (ul) | Inoculum volume from LB (ul) | Volume of M9 added (ml) |
|---|---|---|---|
| pTet luxI cfp(0 ng/ml) | - | 3 ul | 3 ml |
| pTet luxI cfp(1 ng/ml) | 0.3 ul | 3 ul | 3 ml |
| pTet luxI cfp(10 ng/ml) | 3 ul | 3 ul | 3 ml |
| pTet luxI cfp(25 ng/ml) | 7.5 ul | 3 ul | 3 ml |
| pTet luxI cfp(50 ng/ml) | 15 ul | 3 ul | 3 ml |
| pTet luxI cfp(100 ng/ml) | 30 ul | 3 ul | 3 ml |
| K12Z1 (Autoflourescence) | - | 3 ul | 3 ml |
Results: The mean fluorescence of CFP was seen to increase with increasing concentrations of aTc.
- Open Loop Trial
E. coli cells containing pTet.luxI.cells were first cultured in Luria Bertani medium for 10 hours following which they were transferred to M9 medium where they were induced with aTc (50 ng/ml). The cells were then allowed to grow in M9 for 12 hours.
The medium (now containing the autoinducer) was filtered and was added to an equal volume of freshly prepared M9 medium containing twice the concentration of glucose. E.coli cells containing pLac.luxR.yfp.pR.cfp (previously grown in LB medium) were inoculated into the M9 medium prepared and were induced with different concentrations of IPTG. Cells were allowed to grow for 12 hours.
- The Scaling Argument
Two sets of experiments would be carried out, where cells would be grown to an optical density of 0.257 & 0.015 respectively in order to check for density dependence of autoinducer production. The supernatent would be filtered out and the one from the OD=0.257 would be scaled down to OD=0.015. The cells would then be imaged using a phase contrast microscope. The scaling argument would be proved if we get the same cfp expression from both.

