Imperial/Cell-Free/Whatis
From 2007.igem.org
(→What is Cell=Free?) |
|||
| (16 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:IC07navmenu}} | {{Template:IC07navmenu}} | ||
__NOTOC__ | __NOTOC__ | ||
| + | <html> | ||
| + | <link rel="stylesheet" href="/igem07/index.php?title=User:Dirkvs/Stylesheets/IC07persist.css&action=raw&ctype=text/css" type="text/css" /> | ||
| + | |||
| + | <div id="tabs"> | ||
| + | <ul> | ||
| + | <li><a class="current" href="https://2007.igem.org/Imperial/Cell-Free/Whatis" title=""><span>What is Cell-Free?</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Comparison" title=""><span>Advantages of CFS</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Contribution" title=""><span>Our Contributions</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Characterisation" title=""><span>Characterisation</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/Imperial/Cell-Free/Applications" title=""><span>CFS Applications</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </div> | ||
| + | <hr /> | ||
| + | <br clear="all"> | ||
| + | </html> | ||
== What is Cell-Free? == | == What is Cell-Free? == | ||
| - | Cell- | + | '''Cell-Free Systems (CFS)''' involve the ''in vitro'' expression of genes into proteins. These systems can serve as a compatible chassis for the various parts and devices from the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts]. |
| + | |||
| + | |||
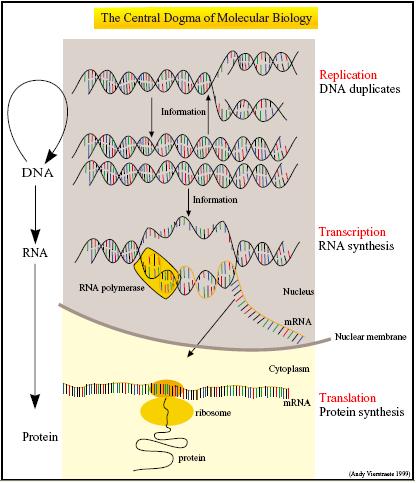
| + | [[Image:IC07_central_dogma.jpg|thumb|300px|left|The Central Dogma (Picture by Andy Verstraete, 1999)]] | ||
| + | '''The Central Dogma''' describes gene expression in terms of two essential processes - the transcription of DNA into messenger RNA (mRNA) and the translation of mRNA into polypeptides.<sup>[[#References |1]]</sup> Not only do CFS house the molecular machinery necessary for transcription and translation, they are also optimized for these two processes. | ||
| + | |||
| + | '''Coupled transcription-translation systems''' usually combine a bacteriophage RNA polymerase and promoter with eukaryotic or prokaryotic extracts rich in ribosomes, transfer RNAs and aminoacyl-tRNA transferase enzymes. Buffers are also added to maintain the appropriate magnesium and salt concentrations required for efficient translation. In addition, an ATP regenerating system involving either creatine phosphate and creatine kinase or phosphoenolpyruvate and pyruvate kinase is used to power and prolong the lifespan of the expression machinery.<sup>[[#References |2]]</sup> | ||
| + | |||
| + | '''A good analogy''' compared this transcription-translation machinery of the CFS to the hardware and the synthetic DNA to the software.<sup>[[#References |3]]</sup> This gives an apt illustration of the ease of building genetically engineered machines using the cell-free approach. Simply by adding the DNA template to the cell extract and feeding solution, the CFS would be able to express the encoded genetic circuit. | ||
| + | |||
| + | <br clear="all"> | ||
| + | |||
| - | + | As a forward step to stricter quality control, as well as the specifications of our projects necessitating a cell-free environment, part of our contributions to iGEM involve the investigation and characterization of cell-free expression systems as a new chassis for the Registry. | |
| Line 12: | Line 39: | ||
|- style="background:#ffffff; text-align:left;" color:white;" align="center" | |- style="background:#ffffff; text-align:left;" color:white;" align="center" | ||
|| [[Image:Icgems-cellextract.png]] | || [[Image:Icgems-cellextract.png]] | ||
| - | |style="padding-left:30px;"|''In vitro'' synthesis of proteins using cell-free extracts consists of two main processes - '''transcription''' of DNA into messenger RNA (mRNA) and '''translation''' of mRNA into polypeptides. | + | |style="padding-left:30px;"|''In vitro'' synthesis of proteins using cell-free extracts consists of two main processes - '''transcription''' of DNA into messenger RNA (mRNA) and '''translation''' of mRNA into polypeptides. In addition to the coupled transcription-translation systems described above, the recently developed [http://www.nature.com/nbt/journal/v19/n8/full/nbt0801_732.html PURE] system is a reconstituted CFS that synthesizes proteins using recombinant elements.<sup>[[#References |4]]</sup> This purely synthetic expression system enables even better quality control over the reaction conditions. |
| + | |||
|} | |} | ||
<br clear="all"> | <br clear="all"> | ||
| Line 40: | Line 68: | ||
|} | |} | ||
<br clear="all"> | <br clear="all"> | ||
| - | |||
=== Types of Compartmentalization === | === Types of Compartmentalization === | ||
| Line 47: | Line 74: | ||
{| style="width:800px;" cellpadding="2" cellspacing="0" align="center" | {| style="width:800px;" cellpadding="2" cellspacing="0" align="center" | ||
|- style="background:#ffffff; text-align:left;" color:white;" align="center" | |- style="background:#ffffff; text-align:left;" color:white;" align="center" | ||
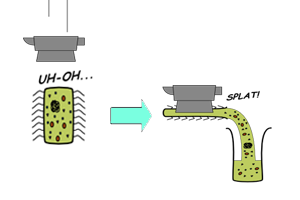
| - | || [[Image:Icgems-bulksol.png| | + | || [[Image:Icgems-bulksol.png|180px]] |
| - | || '''Batch-mode CFS''' | + | || |
| + | '''Batch-mode CFS''' | ||
*Transcription-translation reaction is carried out in bulk solution. Expression is usually limited by nutrient (nucleotides and amino acids) and energy supplies. | *Transcription-translation reaction is carried out in bulk solution. Expression is usually limited by nutrient (nucleotides and amino acids) and energy supplies. | ||
|- style="background:#ffffff; text-align:left;" color:white;" align="center" | |- style="background:#ffffff; text-align:left;" color:white;" align="center" | ||
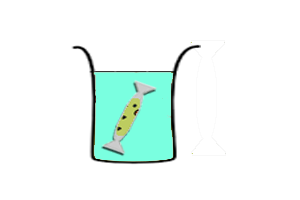
| - | || [[Image:Icgems-exchange.png| | + | || [[Image:Icgems-exchange.png|180px]] |
| - | || '''Continuous-exchange CFS''' | + | || |
| + | '''Continuous-exchange CFS''' | ||
*Transcription-translation reaction is separated from feeding solution by a dialysis membrane. Expression is sustained by diffusion of nutrients from the feeding soltuion to the reaction. Wastes generated by the reaction is diluted in the feeding solution. | *Transcription-translation reaction is separated from feeding solution by a dialysis membrane. Expression is sustained by diffusion of nutrients from the feeding soltuion to the reaction. Wastes generated by the reaction is diluted in the feeding solution. | ||
|- style="background:#ffffff; text-align:left;" color:white;" align="center" | |- style="background:#ffffff; text-align:left;" color:white;" align="center" | ||
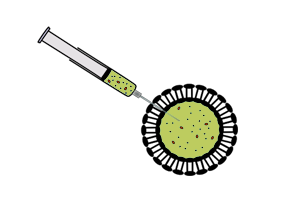
| - | || [[Image:Icgems-vesicles.png| | + | || [[Image:Icgems-vesicles.png|180px]] |
| - | || '''Vesicle-encapsulated CFS''' | + | || |
| - | *The reaction is separated from feeding solution by a phospholipid bilayer. Expression is maintained for a longer time period than batch-mode CFS because of exchange of materials between the reaction and the feeding solution across the membrane. More reliable exchange of materials is established by inserting a non-specific pore protein with a suitable channel size into the phospholipid bilayer. | + | '''Vesicle-encapsulated CFS''' |
| + | *The reaction is separated from feeding solution by a phospholipid bilayer. Expression is maintained for a longer time period than batch-mode CFS because of exchange of materials between the reaction and the feeding solution across the membrane. More reliable exchange of materials is established by inserting a non-specific pore protein with a suitable channel size into the phospholipid bilayer.<sup>[[#References |3]]</sup> | ||
|} | |} | ||
<br clear="all"> | <br clear="all"> | ||
| + | <center> On to next stage: | [https://2007.igem.org/Imperial/Cell-Free/Comparison Advantages of CFS >>]</center> | ||
| - | + | == References == | |
| + | # Crick F. Central dogma of molecular biology. Nature 1970 Aug 8; 227(5258) 561-3. pmid:4913914. | ||
| + | # Pellinen T, Huovinen T, and Karp M. A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal Biochem 2004 Jul 1; 330(1) 52-7. doi:10.1016/j.ab.2004.03.064 pmid:15183761. | ||
| + | # Noireaux V, Bar-Ziv R, Godefroy J, Salman H, and Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol 2005 Sep 15; 2(3) P1-8. doi:10.1088/1478-3975/2/3/P01 pmid:16224117. | ||
| + | # Shimizu Y, Kanamori T, and Ueda T. Protein synthesis by pure translation systems. Methods 2005 Jul; 36(3) 299-304. doi:10.1016/j.ymeth.2005.04.006 pmid:16076456. | ||
Latest revision as of 19:59, 26 October 2007

What is Cell-Free?
Cell-Free Systems (CFS) involve the in vitro expression of genes into proteins. These systems can serve as a compatible chassis for the various parts and devices from the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts].
The Central Dogma describes gene expression in terms of two essential processes - the transcription of DNA into messenger RNA (mRNA) and the translation of mRNA into polypeptides.1 Not only do CFS house the molecular machinery necessary for transcription and translation, they are also optimized for these two processes.
Coupled transcription-translation systems usually combine a bacteriophage RNA polymerase and promoter with eukaryotic or prokaryotic extracts rich in ribosomes, transfer RNAs and aminoacyl-tRNA transferase enzymes. Buffers are also added to maintain the appropriate magnesium and salt concentrations required for efficient translation. In addition, an ATP regenerating system involving either creatine phosphate and creatine kinase or phosphoenolpyruvate and pyruvate kinase is used to power and prolong the lifespan of the expression machinery.2
A good analogy compared this transcription-translation machinery of the CFS to the hardware and the synthetic DNA to the software.3 This gives an apt illustration of the ease of building genetically engineered machines using the cell-free approach. Simply by adding the DNA template to the cell extract and feeding solution, the CFS would be able to express the encoded genetic circuit.
As a forward step to stricter quality control, as well as the specifications of our projects necessitating a cell-free environment, part of our contributions to iGEM involve the investigation and characterization of cell-free expression systems as a new chassis for the Registry.
Types of Extracts
| In vitro synthesis of proteins using cell-free extracts consists of two main processes - transcription of DNA into messenger RNA (mRNA) and translation of mRNA into polypeptides. In addition to the coupled transcription-translation systems described above, the recently developed [http://www.nature.com/nbt/journal/v19/n8/full/nbt0801_732.html PURE] system is a reconstituted CFS that synthesizes proteins using recombinant elements.4 This purely synthetic expression system enables even better quality control over the reaction conditions. |
Comparison between different types of cell extracts
|
|
|
| |
Types of Compartmentalization
Previous research has been done to optimize cell extracts for in vitro protein synthesis. Their endogenous genetic content is removed so that exogenous DNAs or mRNAs can be expressed. Nuclease activity has been reduced and degradation of certain amino acids has been identified. ATP regenerating systems have also been added to improve the energy supply. Different strategies of compartmentalization have been explored to prolong the lifespan of CFS.

|
Batch-mode CFS
|
|
Continuous-exchange CFS
|
|
Vesicle-encapsulated CFS
|
References
- Crick F. Central dogma of molecular biology. Nature 1970 Aug 8; 227(5258) 561-3. pmid:4913914.
- Pellinen T, Huovinen T, and Karp M. A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal Biochem 2004 Jul 1; 330(1) 52-7. doi:10.1016/j.ab.2004.03.064 pmid:15183761.
- Noireaux V, Bar-Ziv R, Godefroy J, Salman H, and Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys Biol 2005 Sep 15; 2(3) P1-8. doi:10.1088/1478-3975/2/3/P01 pmid:16224117.
- Shimizu Y, Kanamori T, and Ueda T. Protein synthesis by pure translation systems. Methods 2005 Jul; 36(3) 299-304. doi:10.1016/j.ymeth.2005.04.006 pmid:16076456.