Imperial/Wet Lab/Protocols/CBD1.2
From 2007.igem.org
(Difference between revisions)
(→Protocols) |
(→Protocols) |
||
| Line 37: | Line 37: | ||
====Protocols==== | ====Protocols==== | ||
#First collect all equipment and reagents and ensure that the fluorometer and that the PC connected has a data collection protocol installed. | #First collect all equipment and reagents and ensure that the fluorometer and that the PC connected has a data collection protocol installed. | ||
| - | #''Commercial E.coli Cell Extract'': First prepare a complete amino acid mixture for both extract solutions: Add the | + | #''Commercial E.coli Cell Extract'': First prepare a complete amino acid mixture for both extract solutions: Add the 10μl volume of two amino acid minus mixtures into an labeled eppendorf to give a volume of 20μl. Each amino acid minus mixture is missing one type of amino acid, and so by combining two solutions we are complementing each solution for the missing amino acid. Place eppendorf in a rack on bench. |
#''Commercial T7 Cell Extract'':Carry out procedure above for the T7 amino acid minus mixtures. | #''Commercial T7 Cell Extract'':Carry out procedure above for the T7 amino acid minus mixtures. | ||
| - | #''Commercial E.coli Cell Extact'':Take a eppendorf tube and add | + | #''Commercial E.coli Cell Extact'':Take a eppendorf tube and add 20µl of the E.coli complete amino acid mixture. Then add add 80µl of S30 Premix Without Amino Acid. Then add 60µl of S30 Extract Circular. |
| + | #Then put 80µl of the pTet | ||
| + | #Finally add nuclease-Free Water to bring final volume (inc.DNA vol) to 60µl, the volume of DNA added will be determined in experiment 1 and the volume of the nuclease free water adjusted accordingly. Place the eppendorf tube in a rack on the bench | ||
#Carry out step above twice more so that we end up with three eppendorf tubes of prepared commercial E.coli extract. | #Carry out step above twice more so that we end up with three eppendorf tubes of prepared commercial E.coli extract. | ||
#''Commercial T7 Cell Extract'':Repeat step 3 and 4 again three times however use the T7 cell extract reagents and the T7 prepare complete amino acid mixture. We will now have three eppendorf tubes of prepared commercial T7 cell extract. | #''Commercial T7 Cell Extract'':Repeat step 3 and 4 again three times however use the T7 cell extract reagents and the T7 prepare complete amino acid mixture. We will now have three eppendorf tubes of prepared commercial T7 cell extract. | ||
Revision as of 20:36, 26 October 2007

Wet Lab: Protocols: Initial Testing for DNA Constructs in vitro
Aims:
- To determine which constructs for cell by date expresses in vitro
- To test if pTet-GFP ([http://partsregistry.org/Part:BBa_I13522 BBa_I13522]) and pT7-GFP ([http://partsregistry.org/Part:BBa_E7104 BBa_E7104]) works in commercially bought S30 E.coli cell extract for circular DNA, and S30 T7 e.coli cell extract for circular DNA respectively.
Equipment
- Fluorometer + PC
- 37°C incubator
- 1 Fluorometer plate (black)
- Sticky seal tape
- Gilson pipettes p200 p20 p10
- Eppendorf Tubes
- Stopwatch
- Foil
Reagents
- Commercial S30 E.coli extract. Including:
- 175µl Amino Acid Mixture Minus Cysteine, 1mM
- 175µl Amino Acid Mixture Minus Methionine, 1mM
- 175µl Amino Acid Mixture Minus Leucine, 1mM
- 450µl S30 Extract, Circular (3 × 150µl)
- 750µl S30 Premix Without Amino Acids
- Commercial S30 T7 extract. Including:
- 175µl Amino Acid Mixture Minus Cysteine, 1mM
- 175µl Amino Acid Mixture Minus Methionine, 1mM
- 175µl Amino Acid Mixture Minus Leucine, 1mM
- 450µl T7 S30 Extract, Circular (3 × 150µl)
- 750µl S30 Premix Without Amino Acids
- Nuclease Free water x1ml
- DNA pTet-GFP from midiprep
- GFP solution (For this initial experiment does not need to be purified GFP, we just want to know we have the right filter and that our settings are adjusted to measuring GFP)
Protocols
- First collect all equipment and reagents and ensure that the fluorometer and that the PC connected has a data collection protocol installed.
- Commercial E.coli Cell Extract: First prepare a complete amino acid mixture for both extract solutions: Add the 10μl volume of two amino acid minus mixtures into an labeled eppendorf to give a volume of 20μl. Each amino acid minus mixture is missing one type of amino acid, and so by combining two solutions we are complementing each solution for the missing amino acid. Place eppendorf in a rack on bench.
- Commercial T7 Cell Extract:Carry out procedure above for the T7 amino acid minus mixtures.
- Commercial E.coli Cell Extact:Take a eppendorf tube and add 20µl of the E.coli complete amino acid mixture. Then add add 80µl of S30 Premix Without Amino Acid. Then add 60µl of S30 Extract Circular.
- Then put 80µl of the pTet
- Finally add nuclease-Free Water to bring final volume (inc.DNA vol) to 60µl, the volume of DNA added will be determined in experiment 1 and the volume of the nuclease free water adjusted accordingly. Place the eppendorf tube in a rack on the bench
- Carry out step above twice more so that we end up with three eppendorf tubes of prepared commercial E.coli extract.
- Commercial T7 Cell Extract:Repeat step 3 and 4 again three times however use the T7 cell extract reagents and the T7 prepare complete amino acid mixture. We will now have three eppendorf tubes of prepared commercial T7 cell extract.
- Vortex the tubes to mix thoroughly and place the 5x eppendorf tubes in the incubator at 37oC
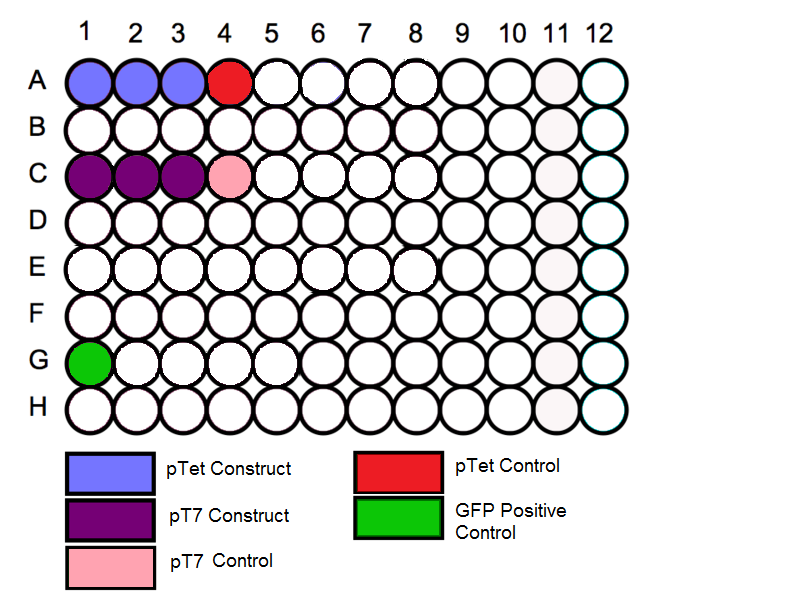
Loading Plate
- Follow the schematic for the plate and begin by loading the in vitro expression system into the correct wells. Before loading in the samples vortex the tubes for a few seconds to mix the solution.
- Place the lid on the 96well plate and tape up the edges of the lid. This should be put into the incubator at 37oC for 10 minutes to allow temperature to equilibrate
- Remove from 37oC incubator and spin-down in centrifuge in plate centrifuge at 2000rpm for a few seconds. Spin down is the process of bringing down any solution on lid or side of well into the base of the well. Alternatively can tap the top of the lid to bring down any solution to bottom of the well.
- Remove lid off th e 96well plate and place in the fluorometer. Create a file name protocol 2-1 under: D:\IGEM\INSERT DATE\CBD\ protocol 2-1. Export the data here. If repeated measurements change the second number to suit repeat number, e.g. 2nd repeat protocol 2-2, 5th repeat protocol 2-5. Once the data collection is set up then initiate the measurements.
- Place the plate in the fluorometer to measure its initial fluorescent reading.
- After the measurement, place the sticky tape across the plate, and put the plate in the 37oC incubator.
- Before placing them in the water bath, wrap aluminium foil around them to prevent photobleaching.
- Repeat the reading every 30 minutes, for 6 hours.
Schematic