Paris/Notebook All
From 2007.igem.org
July 1
July 2
yesterday -- tomorrow
First day in the lab !
Overview of the project
Planning of the lab work
Primer design
Preliminary work on w121 strain
Transduction of DapA- deletion from w121 to MG1655
We got the W121 strain from a lab in Pasteur Institute. This strain is [DapA-; Erythromycin R], but also has a couple of other mutations we are not interested in. Thus we will need to do a transduction of the deletion to the strain we will use: MG1655
We launched ON (= OverNight) culture of w121 to prepare a stock of P1 phages.
Measuring kinetic parameters of our strains
In order to model the dynamics of the synthetic organism, we need to measure several parameters. Two of which are:
- The growth of the [DapA-] strain relative to the DAP concentration of the medium;
- The excretion of DAP by the [DapA+] strain.
The first one is easy, we just need to measure growth kinetics of our [DapA-] strain contingent on the concentration of the DAP we add in the medium. The second one is more tricky. Direct dosage of DAP is quite complicated and expensive, thus we will try a kind of bio-measurement. We will grow the [DapA+] strain ON, then we will centrifugate to recover the culture medium. We will filter this culture medium to sterilize it and we will grow a [DapA-] strain in it. The growth curve should enable us evaluating the medium concentration in DAP.
July 3
Preparation of the P1 stock on the w121 strain.
See protocols. This preparation is necessary for transduction of DapA deletion in MG1655 strain.
Kinetic measurements:
As previously described, we want to measure :
- Growth of DapA- strain (w121) relative to the concentration of DAP in the medium :
- The excretion of Dap by MG1655 DapA+ strain by an indirect way
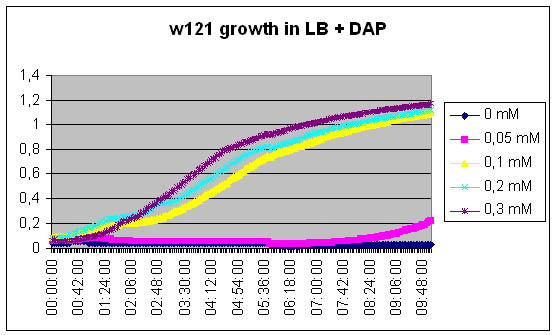
Growth of DapA- strain (w121) relative to the concentration of DAP in the medium
We measure the kinetic of the w121 as a function of DAP concentration
[DAP] = 5mM
| Kinetic Array :Growthing of w121 strain 'supplemented' with S0 (1µL w121 strain incubated ON) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | ||||||||||||
| B | 200µL LB + 0µL DAP | 200µL LB + 2µL DAP | 200µL LB + 4µL DAP | 200µL LB + 8µL DAP | 200µL LB + 12µL DAP | |||||||
| C | ||||||||||||
| D | ||||||||||||
| E | ||||||||||||
| F | ||||||||||||
| G | ||||||||||||
| H | ||||||||||||
See Results.
Measuring excretion of Dap by MG1655 DapA+ strain in an indirect manner
We measure the growth of DapA- strain in the filtered growth medium of previously incubated MG1655 strain (S0).
S0 = Centrifuged and filtered medium of MG1655 ON
[DAP] = 5mM
| Kinetic Array :Growthing of w121 strain 'supplemented' with S0 (1µL w121 strain incubated ON) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | ||||||||||||
| B | 200µL S0 + 0µL DAP | 200µL S0 + 2µL DAP | 200µL S0 + 4µL DAP | 200µL S0 + 8µL DAP | 200µL S0 + 12µL DAP | |||||||
| C | ||||||||||||
| D | ||||||||||||
| E | ||||||||||||
| F | ||||||||||||
| G | ||||||||||||
| H | ||||||||||||
See Results.
Acinetobacter calcoaceticus ADP1
Culture
We received the strain Acinetobacter calcoaceticus ADP1 from Pasteur Institute (Center of biological resources of the Pasteur Institute): code CIP (ATCC number 33305). The strain was dehydrated and cultured following this protocol on LB agar plate at 30°C overnight.
See protocols
Medium to triglyceride/wax esters synthesis
Acinetobacter ADP1 is able to multiply in LB medium at 30°C. It is also able to synthesise lipids (triglyceride and wax ester) under specific conditions: Low Nitrogen Minimum Medium (LNMM).
This medium contains low nitrogen leading the bacterium to decrease its growing. With carbon hydrate source (here succinic acid), the bacteria stock energy into lipids (triglyceride and wax esters).
See protocols
<<home
July 4
Transduction to MG1655 using the P1 stock made on w121.
See protocols.
Growth kinetics : Results of July, 3
Growth of DapA- strain (w121) relative to the concentration of DAP in the medium
Measuring excretion of Dap by MG1655 DapA+ strain in an indirect manner
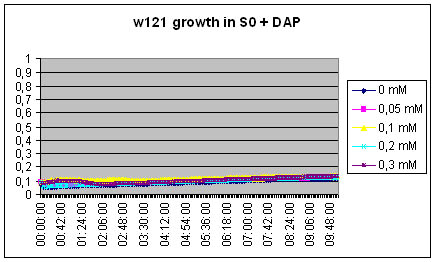
The w121 did not grow at all in the S0 medium... Either we did something wrong, or there is a logical explanation ! There might be growth inhibitors in the ON medium, their might be no nutriments left in the medium...
We will redo the experiment but this time, the medium will be recovered from exponential phase culture of MG1655
Acinetobacter liquid culture
We picked up a colony from LB solid culture from 07/03/2007 and put it into 5mL LB medium (30°C aerobic) (overnight culture).
Low Nitrogen Minimum Medium with Nile Red
This medium allows Acinetobacter ADP1 to synthesize triglyceride and wax ester. Lipid inclusions are seen by fluorescent dye (Nile Red).
See protocols.
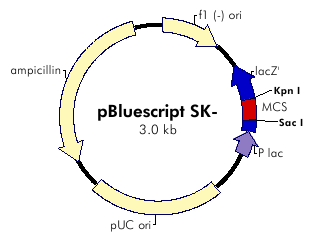
pKS::DGAT Plasmid: transformation
The plasmid contains ampicilline resistance gene and pLac promotor upstream the transgene DGAT (Diacylglycerol Acyltransferase).
We cloned the plasmid by bacterial transformation (Subcloning Efficiency DH5alpha Competent Cells: See protocols).
July 5
Preliminary work on w121 strain
As previously observed, the w121 strain did not grow at all in the S0 medium... We supposed that their might be no nutriments left in the medium because we used a medium in which MG1655 grew ON... We will redo the experiment but this time, the medium will be recovered from exponential phase culture of MG1655
- We started a culture 150 ml of MG1655, and we take samples of the culture at different OD during exponential phase :
- At OD = 0.2 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.2
- At OD = 0.4 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.4
- At OD = 0.6 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.6
- At OD = 0.8 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.8
Transduction of the DapA deletion into MG1655 failed...
We will redo the stock of P1 phages.
Glycerol stock of Acinetobacter ADP1
See protocols
Solid culture of Acinetobacter ADP1
We spread Acinetobacter on agar LNMM with and without Nile Red.
We spread E.Coli DH5alpha transformed by pKS::DGAT or not on agar LNMM with and without Nile Red.
See July6 for the results.
July 6
Preparing growth medium for transduction screening
If the transduction works (i.e. an homologous recombinaison that should delete DapA by inserting an erythromycin resistant cassette), transducted MG1655 Coli should grow in a medium LB+erythro+ DAP but can't grow in a medium LB+erythro without DAP.
- Preparation of DAP solution from the powder (50mM). See Protocols
- Making 10 petri dish (LB+tet+citrate+DAP). See Protocols
- Making 10 petri dish (LB+erythromycin+citrate+DAP). See Protocols
Acinetobacter and E.Coli on LNMM Nile Red solid culture
Nile Red can be excited by light around 312nm (Spiekermann,1999). After 24h incubation, we observed Acinetobacter and E.coli on LNMM with and without Nile Red:
We can observe basic fluorescence with Acinetobacter on LNMM with and without Nile Red. With E.coli transformed or not with pKS::DGAT, we do not observe fluorescence; indeed, coli do not grow on LNMM...
We decided to wait more to see acccumulation of triglyceride with Acinetobacter.
July 7
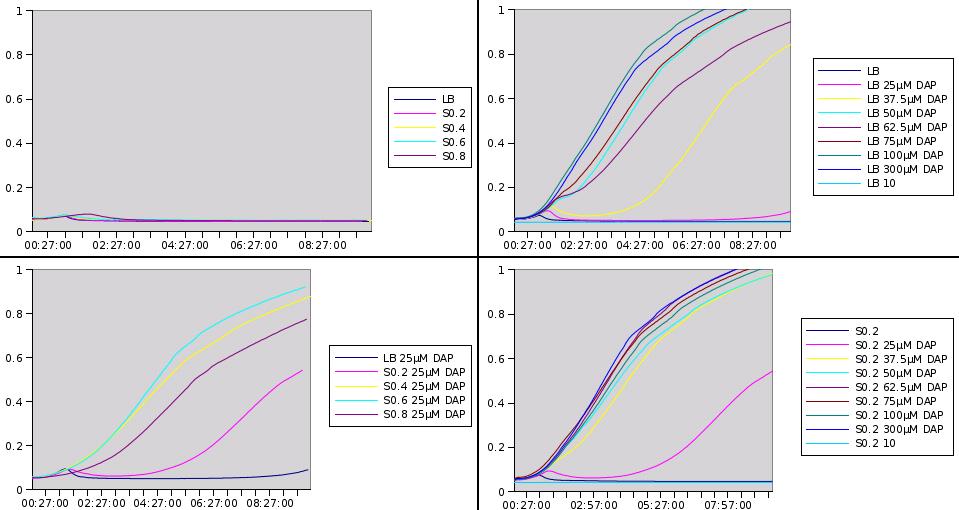
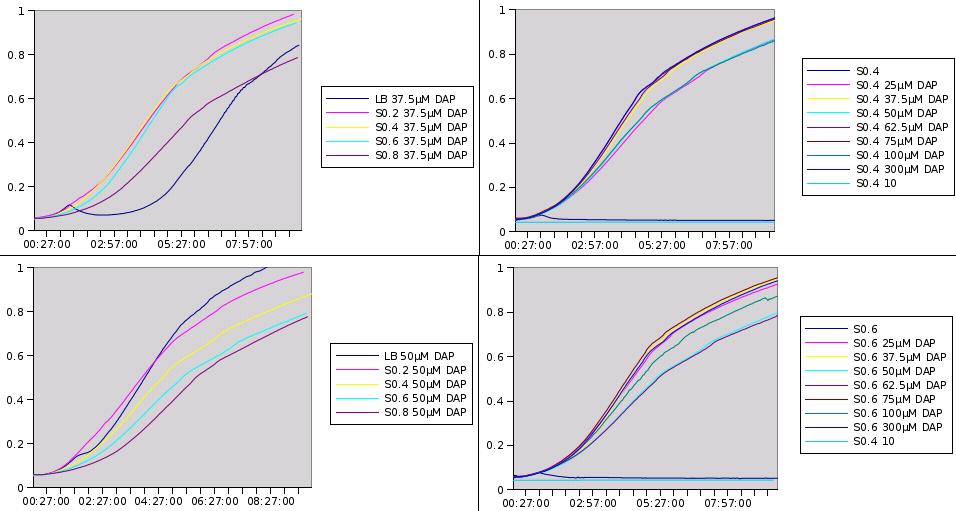
Growth kinetics of w121 strain
We make an array to test growth of w121 on different growth media (LB, S0.2, S0.4, S0.6, S0.8), supplemented with different amounts of DAP.
Two questions are addressed by the following assay:
- What is the growth behaviour of w121 (dapA- strain) at different concentrations of DAP?
- How does w121 strain grow on filtrates of MG1655 growth medium; that is, does MG1655 secrete DAP during growth?
MG1655 was grown on LB medium and the growth medium was filtered free of bacteria at different DO (Optical Densities) during exponential growth phase:
S0.2 (at DO=0.2) S0.4 (at DO=0.4) S0.6 etc S0.8
W121 was grown on different media:
LB line B in the array S0.2 (at DO=0.2) line C in the array S0.4 line D in the array S0.6 line E in the array S0.8 line F in the array
In the different columns, DAP was added to the indicated final concentrations (without taking into account DAP produced by MG1655 regarding the recycled growth media)
| Kinetic Array :w121 kinetic as a function of DAP and supplemented medium (S0.x) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | |
| B | H20 | LB+0µM DAP | LB+25µM DAP | LB+37.5µM DAP | LB+50µM DAP | LB+62.5µM DAP | LB+75µM DAP | LB+100µM DAP | LB+300µM DAP | LB+0µM DAP | H20 | |
| C | H20 | S0.2+0µM DAP | S0.2+25µM DAP | S0.2+37.5µM DAP | S0.2+50µM DAP | S0.2+62.5µM DAP | S0.2+75µM DAP | S0.2+100µM DAP | S0.2+300µM DAP | S0.2+0µM DAP | H20 | |
| D | H20 | S0.4+0µM DAP | S0.4+25µM DAP | S0.4+37.5µM DAP | S0.4+50µM DAP | S0.4+62.5µM DAP | S0.4+75µM DAP | S0.4+100µM DAP | S0.4+300µM DAP | S0.4+0µM DAP | H20 | |
| E | H20 | S0.6+0µM DAP | S0.6+25µM DAP | S0.6+37.5µM DAP | S0.6+50µM DAP | S0.6+62.5µM DAP | S0.6+75µM DAP | S0.6+100µM DAP | S0.6+300µM DAP | S0.6+0µM DAP | H20 | |
| F | H20 | S0.8+0µM DAP | S0.8+25µM DAP | S0.8+37.5µM DAP | S0.8+50µM DAP | S0.8+62.5µM DAP | S0.8+75µM DAP | S0.8+100µM DAP | S0.8+300µM DAP | S0.8+0µM DAP | H20 | |
| G | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | |
| H | ||||||||||||
See results.
The 96 well plate is surrounded by H2O in order to maintain humidity during the assay. In each slot, 200µL of the growth medium (LB or S0.x) is mixed with 2µL of w121 culture grown ON, and with different amount of DAP (see table). The growth profile is measured over a period of 10H, a DO measurement is acquired each 3min.
July 8
Growth kinetics of w121 strain
We make an array to test growth of w121 on different growth media (LB, S0.2, S0.4, S0.6, S0.8), supplemented with different amount of DAP.
Two questions are addressed by the following assay:
- What is the growth behaviour of w121 (dapA- strain) at different concentrations of DAP?
- How does w121 strain grow on filtrates of MG1655 growth medium; that is, does MG1655 secrete DAP during growth?
MG1655 was grown on LB medium and the growth medium was filtered free of bacteria at different DO (Optical Densities) during exponential growth phase:
S0.2 (at DO=0.2) S0.4 S0.6 S0.8
W121 was grown on different media:
LB line B in the array S0.2 (at DO=0.2) line C in the array S0.4 line D & E in the array S0.6 line F in the array S0.8 line G in the array
In the different columns, DAP was added to the indicated final concentrations (without taking into account DAP produced by MG1655 regarding the recycled growth media)
| Kinetic Array :w121 kinetic as a function of DAP and supplemented medium (S0.x) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 |
| B | H20 | LB+0µM DAP | LB+13µM DAP | LB+17µM DAP | LB+20µM DAP | LB+23µM DAP | LB+27µM DAP | LB+30µM DAP | LB+33µM DAP | LB+37µM DAP | LB+40µM DAP | H20 |
| C | H20 | S0.2+0µM DAP | S0.2+13µM DAP | S0.2+17µM DAP | S0.2+20µM DAP | S0.2+23µM DAP | S0.2+27µM DAP | S0.2+30µM DAP | S0.2+33µM DAP | S0.2+37µM DAP | S0.2+40µM DAP | H20 |
| D | H20 | S0.4+0µM DAP | S0.4+13µM DAP | S0.4+17µM DAP | S0.4+20µM DAP | S0.4+23µM DAP | S0.4+27µM DAP | S0.4+30µM DAP | S0.4+33µM DAP | S0.4+37µM DAP | S0.4+40µM DAP | H20 |
| E | H20 | S0.4+0µM DAP | S0.4+13µM DAP | S0.4+17µM DAP | S0.4+20µM DAP | S0.4+23µM DAP | S0.4+27µM DAP | S0.4+30µM DAP | S0.4+33µM DAP | S0.4+37µM DAP | S0.4+40µM DAP | H20 |
| F | H20 | S0.6+0µM DAP | S0.6+13µM DAP | S0.6+17µM DAP | S0.6+20µM DAP | S0.6+23µM DAP | S0.6+27µM DAP | S0.6+30µM DAP | S0.6+33µM DAP | S0.6+37µM DAP | S0.6+40µM DAP | H20 |
| G | H20 | S0.8+0µM DAP | S0.8+13µM DAP | S0.8+17µM DAP | S0.8+20µM DAP | S0.8+23µM DAP | S0.8+27µM DAP | S0.8+30µM DAP | S0.8+33µM DAP | S0.8+37µM DAP | S0.8+40µM DAP | H20 |
| H | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 |
The 96 well plate is surrounded by H2O in order to maintain humidity during the assay. In each slot, 200µL of the growth medium (LB or S0.x) is mixed with 2µL of w121 culture grown ON, and with different amount of DAP (see table).
The growth profile is measured over a period of 20H, a DO measurement is acquired each 4min10s.
Results of previous day kinetics
July 9
Miniprep pKS::DGAT
We wanted to clone the plasmid pKS::DGAT after overnight culture (LB-ampicilline) of DH5alpha transformed with pKS::DGAT. See Protocols.
July 10
yesterday -- tomorrow
Nile Red can be excited by light around 312nm (Spiekermann,1999). After 72h incubation, we observed Acinetobacter on LNMM with and without Nile Red:
The negative control is at 24h after incubation:
July 11
Preparation of LB agar plate for E.coli transformed by pKS::DGAT
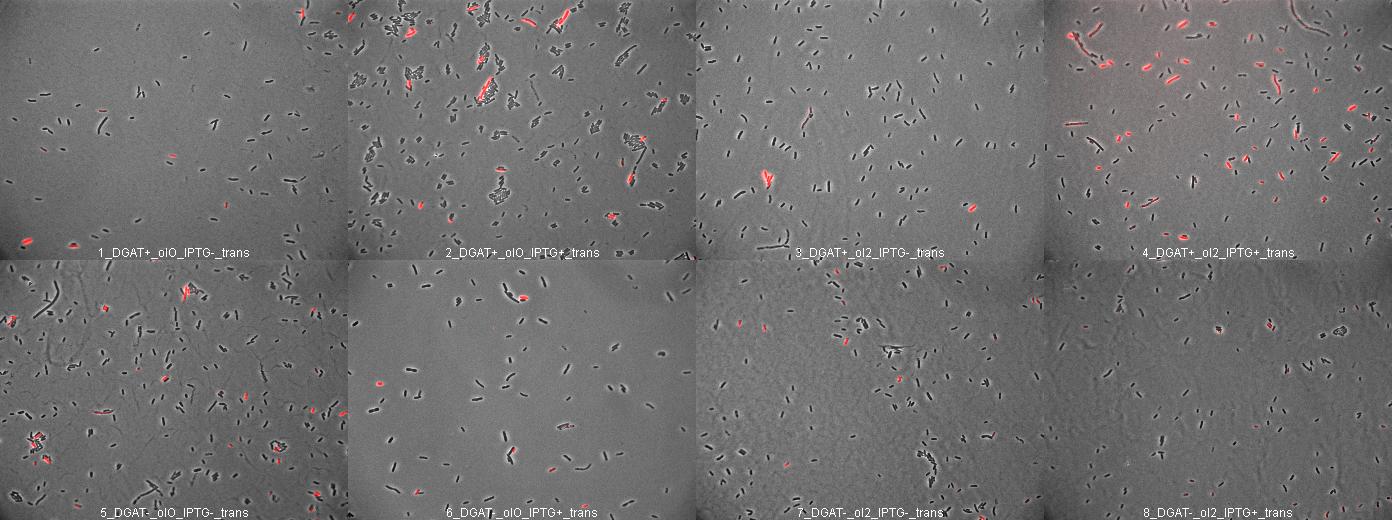
We decided to test the synthesis of triglyceride by E.coli transformed with pKS::DGAT on LB culture. Knowing that when bacteria are in presence of long chain fatty acid (oleate for example), fatty acid are transported into the cytoplasm by fadL (fatty acid degradation), are added acetyl-CoA by fadD and then the fatty acyl-coA is able to bind fadR and repress it. FadR is a repressor of the expression of fadL (the transporter) and the degradation machinery. Culture in presence of long chain fatty acid enhances the transport of fatty acid into the cytoplasm to be dregraded by beta-oxidation. We would like to use this enhanced transport to synthesize triglyceride by DGAT (substrate: long chain fatty acil-CoA and diacylglycerol).
We prepared different medium:
- LB agar + ampicillin (100µg/mL) +/- IPTG (0.4mM) +/- sodium oleate (0.5mM and 2mM) + Nile red (0.5µg/mL of medium). - Minimum medium + ampicillin (100µg/mL) +/- IPTG (0.4mM) +/- sodium oleate (0.5mM and 2mM) + Nile red (0.5µg/mL of medium).
We spread DH5alpha pKS::DGAT (liquid culture from 07/04/2007).
July 12
Preparation of a stock of phage on w121:
See Protocols
Titration of bacteriophages P1
Nicolas C.
We measure the strength of former P1 preparation :
- The stock prepared the 3.7.7
- The stock prepared the 8.7.7
- The stock prepared today (12.7.7)
See results.
See protocols for details.
Culture of FtsZ TS
David B
We want to isolate a good clone of the strain FstZ TS
From the plate of FtsZ TS84 121:
isolation of 6 clones of the 121.1 on LBA+tet incubated at 42°C + Culture ON at 30°C
If one clone do not grow at 42, we'll use it to do the transduction.
See result.
Acinetobacter microscopy
We looked under 5X fluorescence microscopy the fluorescence of colonies of Acinetobacter incubated on LNMM Nile Red for 4-5 days:
Visualisation of colonies stained by Nile Red. It seems that only cells in the middle of colonies (i.e. "old" cells) are producing triglycerides whereas cells on the edges doesn't. Without Nile red, the colonies show no fluorescence.
E.coli pKS::DGAT on LB oleate medium
We incubated E.coli pKS::DGAT overnight on different LB medium (see July 11).
We did not observe any colony. Probably because spread plates with liquid culture kept in the freezer for one week was not a good idea...
On minimum medium, it did not grow too because what we thought to be M9 medium was only agar...
July 13
Test of Ftsz TS strain
The isolated clones of FtsZ TS all grew at 42°C and at 30°C. They must be mutants... So next time we'll try to find other clones from the source of the strain.
Transduction of MG1655 with P1 stock made on w121
We used 03/07/07 (I) and 12/07/07 (II) phage stock from W121 and MG1655 culture overnight.
- Control (1mL LB MgSO4 30mM; CaCl2 15mM)
- 5µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight
- 50µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight
- 500µL Phage + 500µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight
See Protocols
Transformation of Biobricks from the iGEM2007 plates into DH5aplha
- BBa_I0500: Inducible pBad/araC in pSB2K3 (KanR)
well: 9I, Plate 2
- BBa_J23100: strong constitutive promoter in BBa_J61002 (AmpR)
well: 21E, Plate 3
- BBa_B0015: double terminator (B0010-B0012) in pSB1AK3 (AmpR)
well: 1I, Plate 1
Unfreeze chemio-competent bacteria (E.coli DH5alpha; Invitrogen) on ice 5 min
Add 1µL of each resuspended plasmid of interest into 50µL of precedent competent bacteria on ice
Incubate on ice 30 min
heat Shock cells for 20 sec in a 42°C water bath without shaking
Place tube on ice for 2 min
Add 950µL of pre-waemed medium of choice to each tube
Incubate tubes at 37°C for 1 hour at 200 rpm
Spread 20 and 200µL from each transformation on pre-warmed selective plates.
- BBa_I0500: Inducible pBad/araC on KanR (100µg/mL) LB-agar
- BBa_J23100: strong constitutive promoter on AmpR (100µg/mL) LB-agar
- BBa_B0015: double terminator (B0010-B0012) on AmpR(100µg/mL) LB-agar
Incubate plates ON at 37°C
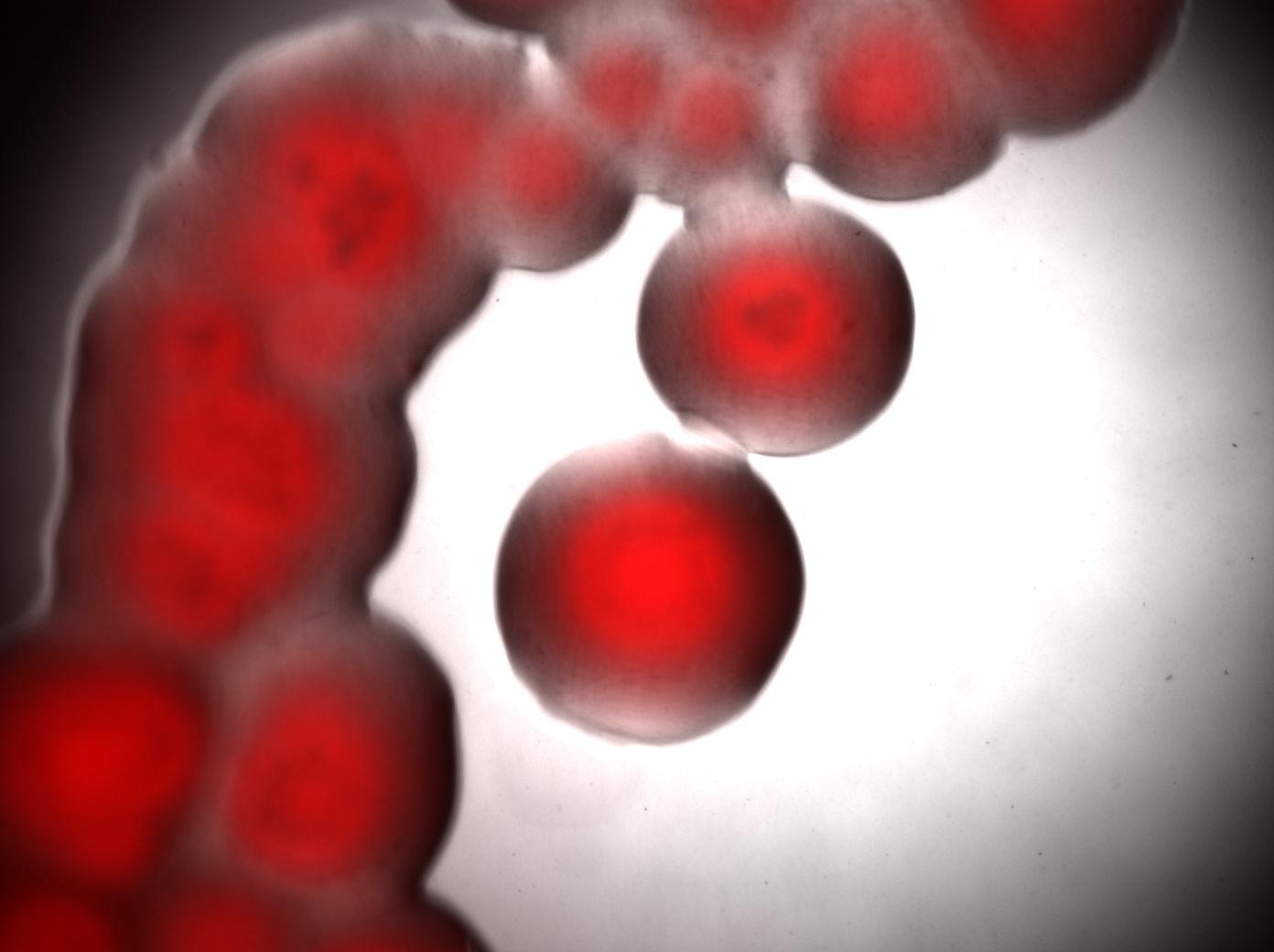
Microscopy of Acinobacter strain
We learn how to use the microscope to see single cells (see Protocols).
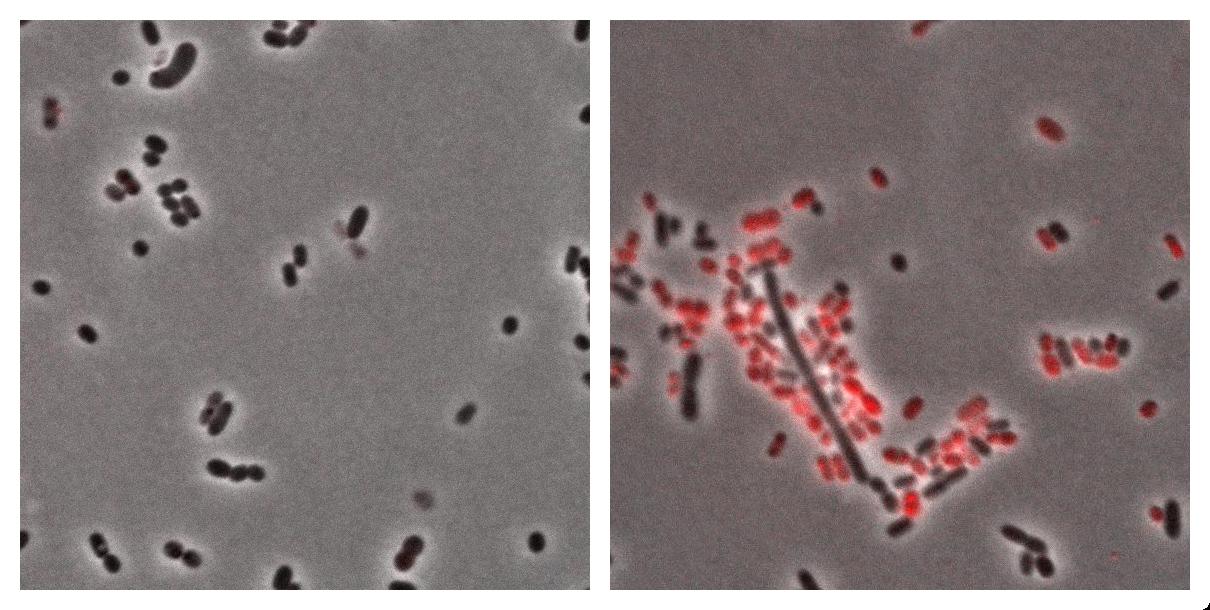
Here we show the photos taken from Acinobacter cells
- left : Acinetobacter grown in Low Nitrogen Mineral medium.
- right : Acinetobacter grow in Low Nitrogen Mineral medium with Nile Red .
Exposure time and scaling is the same for both images. Clearly, some cells on the right are stained by Nile Red, which is specific to triglycerides, whereas almost all cells on the left are not stained. Tipically, the size of one cell is about 5µm.
Bottom image : visualisation of colonies stained by Nile Red. It seems that only cells in the middle of colonies (i.e. "old" cells) are producing triglycerides whereas cells on the edges doesn't.
PCRs
We received the oligos !!! But not all of them :-(
3 PCRs:
- Lox71-FtsA-FtsZ1 : 3 (Lox71-FtsA-F) + 4 (DEcoR1-FtsZ-R)
- FtsZ2: 5 (DEcoR1-FtsZ-F) + 2 FtsZ-R
- Lox66-DapAColi: 6 (Lox66-DapAColi-F) + 7 (DapAColi-R)
Titration of bacteriophages P1
We measure the strength of former P1 preparation :
- 3.7.7 : ~105/ml : very bad
- 8.7.7 : ~102/ml : very bad
- 12.7.7 : pb : we had the phages too late, and we can't conclude.
July 14
yesterday -- tomorrow
Bastille's Day!!!
What to do ?
- Remove the plate of the transduction experiment and the transformation ones from the 37°C incubator (Upper-left shelf): If it has worked and if someones comes on Sunday:
- isolation of the clones from the transduction on petri-dishes (LB+DAP+Erm+Citrate)
- LB + antibio culture of the transformants (to make a glycerol stock and to do MiniPreps)
- Make a gel (0.8%) and migrate the PCR products
- Purify them
- Do the assembly PCR
If enough time (we can also wait to have the plasmids to do everything at the same time):
- Digestion of the purifications products with appropriate enzymes
- Purification
What has been done
- Several colonies of transduced MG1655 are obtained (transduction from dapA- strain w121):
- Isolation of 10 different colonies on LB Citrate Erythro DAP plates
- Seeding LB-Amp cultures of the following strains:
- DH5alpha transformed with the plasmid carrying the Biobrick pJ23100
- DH5alpha transformed with the plasmid carrying the Biobrick B0015
- These two overnight cultures will allow us to perform (tomorrow):
- Minipreps to isolate the plasmids carrying the biobricks
- Glycerol stocks of the transformed strains
DGAT expressing E.coli
After transformation of DH5alpha E.coli with pKS::DGAT plasmid, a transformant clone was grown overnight in LB Ampicilline medium.
The culture (using a toothpick) was deposited on several solid LB media for growth:
+/- IPTG (inducer of DGAT expression, dgat gene being carried by pKS::DGAT plasmid)
+/- Nile Red (fat detection dye)
+/- oleate at 2 different concentrations (0.5 and 2mM)
PCRs
These two first PCRs aims at removing the Pst1 site in DGAT gene.
July 15
- Minipreps on the following ON cultures:
- clones 1 & 2 of DH5alpha transformed with biobrick pJ23100
- clones 1 & 2 of ... BB B0015
- Isolation of 10 colonies of the following transformants:
- BBa_pJ23107 (ON culture on LB Amp is launched for clones 1 & 2)
- BBa_I0500 (ON cultures on LB-Kan clones 1 & 2)
Growth kinetics of w121 strain
We make an array to test growth of w121 on different growth media (LB, S0.2, S0.4, S0.6, S0.8), supplemented with different amount of DAP.
Two questions are addressed by the following assay:
1) What is the growth behaviour of w121 (dapA- strain) at different concentrations of DAP?
2) How does w121 strain grow on filtrates of MG1655 growth medium; that is, does MG1655 secrete DAP during growth?
MG1655 was grown on LB medium and the growth medium was filtered free of bacteria at different DO (Optical Densities) during exponential growth phase:
S0.2 (at DO=0.2) S0.4 (at DO=0.4) S0.6 S0.8
W121 was grown on different media & Growth kinetics were measured:
LB line B in the array S0.2 (at DO=0.2) line C in the array S0.4 line D in the array S0.6 line E & F in the array S0.8 line G in the array
In the different columns, DAP was added to the indicated final concentrations (without taking into account DAP produced by MG1655 regarding the recycled growth media)
| Kinetic Array :w121 kinetic as a function of DAP and supplemented medium (S0.x) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 |
| B | H20 | LB+0µM DAP | LB+20µM DAP | LB+25µM DAP | LB+30µM DAP | LB+35µM DAP | LB+40µM DAP | LB+45µM DAP | LB+50µM DAP | LB+55µM DAP | LB+60µM DAP | H20 |
| C | H20 | S0.2+0µM DAP | S0.2+5µM DAP | S0.2+10µM DAP | S0.2+15µM DAP | S0.2+20µM DAP | S0.2+25µM DAP | S0.2+30µM DAP | S0.2+35µM DAP | S0.2+40µM DAP | S0.2+45µM DAP | H20 |
| D | H20 | S0.4+0µM DAP | S0.4+5µM DAP | S0.4+10µM DAP | S0.4+15µM DAP | S0.4+20µM DAP | S0.4+25µM DAP | S0.4+30µM DAP | S0.4+35µM DAP | S0.4+40µM DAP | S0.4+45µM DAP | H20 |
| E | H20 | S0.6+0µM DAP | S0.6+2µM DAP | S0.6+5µM DAP | S0.6+8µM DAP | S0.6+11µM DAP | S0.6+14µM DAP | S0.6+17µM DAP | S0.6+20µM DAP | S0.6+23µM DAP | S0.6+26µM DAP | H20 |
| F | H20 | S0.6+0µM DAP | S0.6+2µM DAP | S0.6+5µM DAP | S0.6+8µM DAP | S0.6+11µM DAP | S0.6+14µM DAP | S0.6+17µM DAP | S0.6+20µM DAP | S0.6+23µM DAP | S0.6+26µM DAP | H20 |
| G | H20 | S0.8+0µM DAP | S0.8+2µM DAP | S0.8+5µM DAP | S0.8+8µM DAP | S0.8+11µM DAP | S0.8+14µM DAP | S0.8+17µM DAP | S0.8+20µM DAP | S0.8+23µM DAP | S0.8+26µM DAP | H20 |
| H | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 | H20 |
See results.
The 96 well plate is surrounded by H2O in order to maintain humidity during the assay. In each slot, 200µL of the growth medium (LB or S0.x) is mixed with 2µL of w121 culture grown ON, and with different amount of DAP (see table). The growth profile is measured over a period of 30H, a DO measurement is acquired each 6min10s.
Gel extraction of PCR products
- PCR products of 13/07/07 & 14/07/07
migration on 1% Agarose gel extraction of bands of interesst with gel extraction Wizard kit according to the manufacturer's instructions
July 16
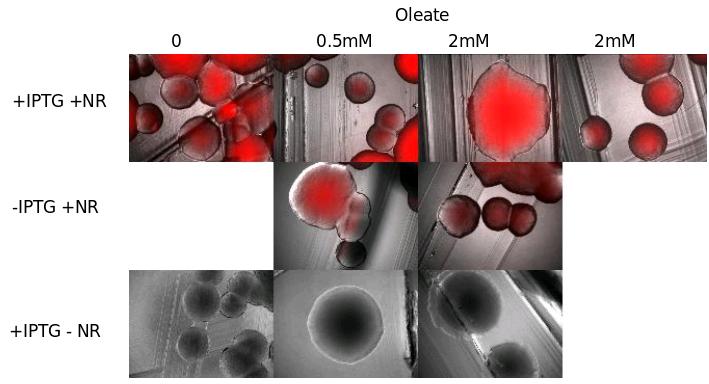
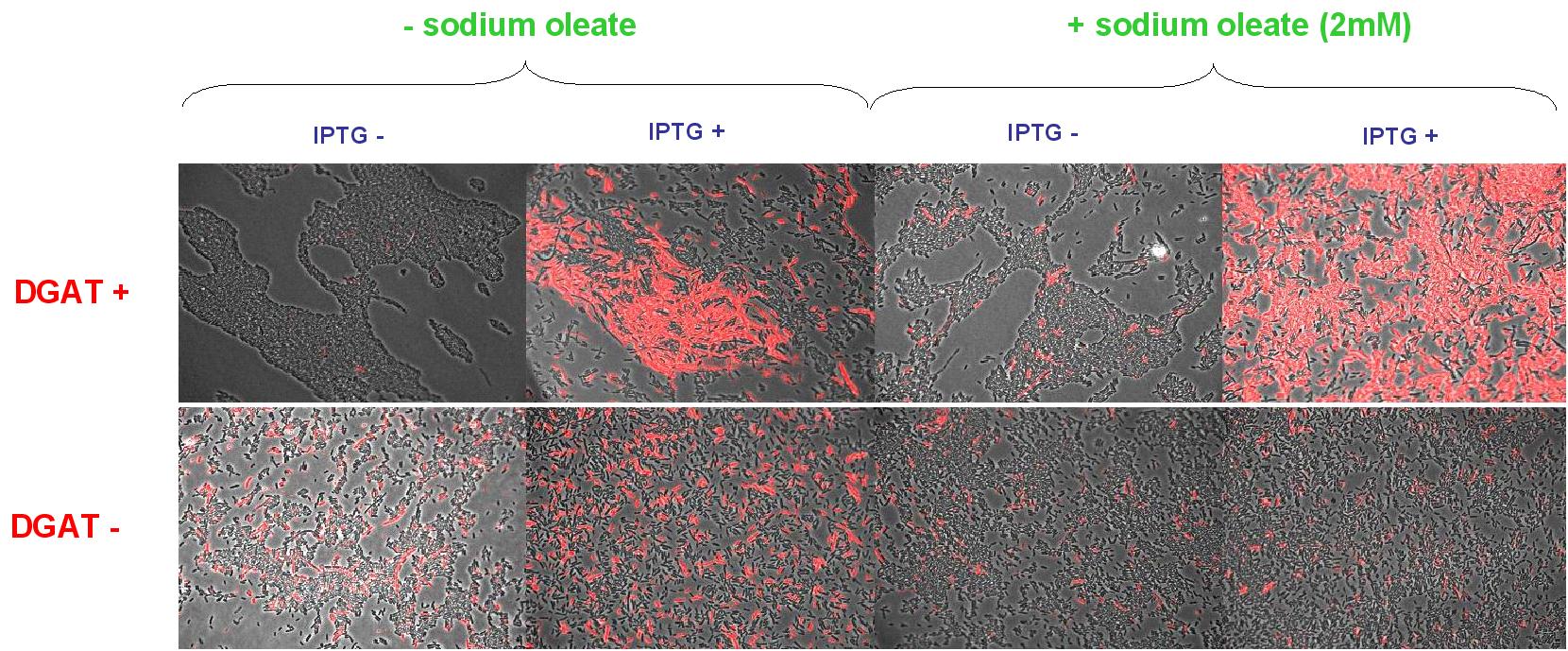
Plasmid pKs::DGAT expression in E. Coli
We tried to see TG with NR died on E. Coli cells transfected with pKs::DGAT, an IPTG-inducible promoter (see July 14) . We tried different growth media containing more or less oleate (0.5 and 2mM), that should in theory increase TG synthesis.
- Observations:
- Interpretation:
We don't see the inducible effect of IPTG.
We can think that :
- Either the fluorescence without IPTG is due to a leak of the promoter.
- Either DGAT is not induce in presence of IPTG, and the fluorescence we see is only a background.
- Perspectives:
- To explain the lack of the promoter pLac, we can notice that we are here in stationary phase, the bacterium metabolism can also change and the presence of lactose derepress LacI leading to expression of DGAT without adding IPTG in the medium.
We also decided to test synthesis of TG in exponential phase.
- To see if the fluorescence is only background, we can use as control E.coli non-transformed by pKS::DGAT.
- We can also wait to see accumulation of TG. Indeed in Acinetobacter, the process is pretty long (2-4 days). It could be the same for E.coli transformed by pKS::DGAT.
We decided to incubate E.coli DGAT and non transformed by pKS::DGAT (transformed by part B0015, plasmid with ampiR) in LB ampicillin medium overnight.
Growth kinetics of w121 strain
Results of the previous day : we lost everything because of a crash of the computer :( Sorry Eimad !
Transduction of MG1655 with P1 stock made on w121
- Control (1mL LB MgSO4 30mM; CaCl2 15mM)
- 5µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ON
- 50µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ON
- 500µL Phage + 500µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ON
=>For the nth time, it is not working : we have only contaminants.
MiniPreps
- I0500 clones 1, 2
- pJ23107 clones 1, 2
PCR purification
- Lox71-FtsZ1
- FtsZ2
- DGAT1
- DGAT2
PCRs
The Lox66-DapAColi PCR did not work... We'll try again with different annealing temperature
Assembly PCRs:
- Lox71-FtsA-FtsZ-1 + FtsZ-2
- DGAT-1 + DGAT-2
The Lox66-DapAColi PCR did not work... so we try a gradient of annealing temperatures
Transformations of Biobricks
As in the protocol given in the registry. Transformation of biobricks :
- BBa_B0030 pSB1A2 : RBS (well 3G plate 1)
- BBa_E0422 pSB1A2 : ECFP (RBS+LVA+Term) (well 11G plate 1)
- BBa_E0241 pSB1A2 : PoPs to GFP converter (well 15c plate 2)
- BBa_E0840 gfp tri-part; strong rbs (well 16E plate 1)
- BBa_J61047 pSB1A2 : Cre ORF (well 8P plate 4)
Transformation in DH5alpha subcloning efficiency
Spread on LB-Amp
July 17
PCRs
- The assembly PCRs did not work... we'll try them again !
- The Lox66-DapAColi PCRs worked very well in all conditions
=> Gel purification
I make a gel to check PCR products. To put on gel :
- 50µL of PCR product
- 5µL of loading buffer.
As the precedent one didn't work, we decided to change our way to do it. We made the PCR reactions within two main steps :
- the 2 PCR products are used as a matrix and added to the PCR mix without the primers ! - then 15 typical cycles
- We add the primers to the PCR mix and run the total for 30 cycles.
We tested the PCR construction. It didn't give anything !
- DGAT = a smear + 1 band at 100kb (primers)
- FtsZ = nothing at all...
Let's do it again tomorow i order to find out where the problem could be from.
DAP solution contamination test
We spread on LB plates DAP solution :
- 50µL of H20 (control)
- 50µL of aliquot DAP 50mM (in use)
- 50µL of DAP 1mM
- 50µL of DAP 0.2 mM
- 50µL of DAP 50mM
+ filtration of the DAP stock
See Results.
Transduction of MG1655 with P1 stock made on w121
As it still didn't work, we try for now with culture of MG1655 in exponential growth rate (ExpGR)
OD=0.6
- Preparation of samples:
- Control (900µL LB MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ExpGR
- 5µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ExpGR
- 50µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ExpGR
- 500µL Phage + 500µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ExpGR
- 20mn @ 37°C = adsorption
- Centrifugation 1mn @ 10000rpm
- Resuspension in LB + Na Citrate 20mM + DAP 300µM
- 1H growth @ 37°C
- Plate on LB + Na Citrate + Erythromycin + DAP
Transformations of Biobricks : results
Transformation of biobricks : we obtained hundreds of clones for each biobrick.
Transformation in DH5alpha subcloning efficiency
Spread on LB-Amp
- BBa_B0030 pSB1A2 : RBS (well 3G plate 1)
- BBa_E0422 pSB1A2 : ECFP (RBS+LVA+Term) (well 11G plate 1)
- BBa_E0241 pSB1A2 : PoPs to GFP converter (well 15c plate 2)
- BBa_E0840 gfp tri-part; strong rbs (well 16E plate 1)
- BBa_J61047 pSB1A2 : Cre ORF (well 8P plate 4)
Transduction of w121 strain
Titration of the stock of phage from 12.7.7.
See results.
Digestions
We did as follow :
- 25µL of plasmid miniprep
- 15,5µL H20
- 5µL of NE2 Buffer 10X (Biolabs)
- 0,5µL of BSA 100X
- 2µL of Enzyme1 (Biolabs)
- 2µL of Enzyme2 (Biolabs)
We have 2 clones for each plasmid, we digested both as follow :
- pJ23100 with EcoR1/Spe1 and Spe1/Pst1
- pJ23107 with EcoR1/Spe1 and Spe1/Pst1
- I0500 with Ecor1/Pst1
- B0015 with EcoR1/Pst1
- Lox66 DapA E.coli (PCR product) with EcoR1/Pst1 and Xba1/Pst1
After 3 hours of digestion, running on a 1% agarose gel.
We cut the bands and put them all at -20°C for next morning.
Exponential culture of E.coli pKS::DGAT and microscopy
- Protocol
- dilution 1/200 of the liquid culture overnight of E.coli pKS::DGAT and E.coli control (see July 16).
- incubation 2hours 37°C
- add:
- oleate : 0/0.5mM/2mM
- IPTG : 0/0.5mM
- incubate 1 hour 37°C
- add Nile Red (0.5µg/mL medium)
- microscopy pictures
- Result:
No differences between the different culture condition (+-IPTG / +-oleate / E.coli transformed or not).
We decide to test after overnight culture: no result.
July 18
Transduction of w121 strain
Titration of the stock of phage from 12.7.7. : ~109 phages/ml : OK
FtsZTS strain screening
We want to test these strains :
- 121.4
- 121.5
- 121.6
- 129
- 1.129
- DCR 14.1
- DCR 14.2
- DCR 14.3
I made glycerol stock of these strains, stored in the freezer. Test :
- Spread these strains on two plates (preheated before spreading) :
- One at 30°C
- One at 42°C
The results tomorrow.
Minipreps
We have 2 clones for each plasmid :
- E0422
- E0241
- E0840
- B0030
- J61047
Elution within 50µL H2O
DNA is within the Miniprep box in the -20°C fridge.
Digestion products
Photos !!!
DAP solution contamination test
Results :
- 50µL of H20 (control) OK
- 50µL of aliquot DAP 50mM (in use) OK
- 50µL of DAP 1mM CONTAMINATED
- 50µL of DAP 0.2 mM OK
- 50µL of DAP 50mM OK
PCRs
| PCR : Assembly PCR Lox71-Fts1-FTsZ1 + FtsZ2 | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2580 | |||
| 50, 55, 60, 65°C | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | NO | ||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 30µL | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Lox71-FtsA-FtsZ1 2µL + FtsZ2 2µL | |||||
E.coli pKS::DGAT
- After overnight culture (see July 17), we do not observe significant triglyceride synthesis.
- We decide to test in solid culture and wait some days. E.coli transformed by pKS::DGAT and the negative control (E.coli transformed by part B0015) were spread on LB medium +- oleate 2mM +- IPTG (0.4mM).
- We try in M9 Minimum Medium (see Protocols) replacing lactose/galactose by 2mM oleate, too.
July 19
FtsZTS strain screening
We want to test these strains :
- 121.4
- 121.5
- 121.6
- 129
- 1.129
- DCR 14.1
- DCR 14.2
- DCR 14.3
I made glycerol stock of these strains, stored in the freezer. Test :
- Spread these strains on two plates (preheated before spreading) :
- One at 30°C
- One at 42°C
Results : every strain grew at 30 and 42°C => FAIL
Overnight Bacterial cultures
We launched several cultures :
- MG1655 -> to do transductions tomorrow (don't forget the DAP, David!)
- pJ23107_J61002 clones 3 & 4 in LB-Amp liquid; also striping clones 3-18 on LB-Amp Agar plates
- I0500_pSB2K3 clones 1 & 2 in LB-Kan liquid medium
- B0015_pSB1AK3 clones 3 & 4 in LB-Kan liquid medium
Tomorrow (20/07/07): perform transduction (MG1655), minipreps, glycerol stocks
July 20
PCRs
| PCR : Lox71-KmR-Lox66 | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 1084 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 10 Lox71-KmR-F | 2.5µL | ? | ||
| 3m00' | Oligo R 10µM | 11 Lox66-KmR-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | MP3_pSB1AK3-BBa_B0015 0.5µl | |||||
| PCR : Lox71-FtsA-FtsZ | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2588 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | ? The PCR seemed to have worked but the purification failed | ||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | toothpick in MG1655 glycerol | |||||
Glycerol stock
- BBa_pJ23107 clone 3&4 (S14)
- BBa_I0500 clone 1&2 (S13)
Minipreps on Biobricks
- BBa_pJ23107 clone 3&4
- BBa_I0500 clone 1&2
E.Coli pKS::DGAT
We look under microscopy 36 hours after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See July 18).
- Observations:
We can observe colonies (X10).
- Interpretation
We can observe lipid inclusions within colonies with DGAT. Further investigations need single cell visualisation.
July 21
Digestion reactions
Double digestion reactions were performed as follows: for a double digestion by X & Y, the template was digested by X(mix1:in a 25µL reaction mix)& Y (mix2: in a 25µL reaction mix) for 2h at 37°. Then, enzyme Y was added to mix1 & enz. X added to mix 2. This second reaction was also performed at 37° for 2h.
25µL reaction mix:
- 10µL DNA template (miniprep or PCR product)
- 1µL enz. (added last)
- 2.5µL 10x reaction buffer (depending on enzymes used)
- BSA 100x 0.25µL
- H2O qsp 25µL
| Digestion Products | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number | Product Name | Matrix Name | Enzyme 1 | Enzyme 2 | size | Description | ||
| D9 | pJ23100 ES vector | pJ23100 (MP1.1 & MP1.2) | EcoI | SpeI | 2096 | Vector with mRFP RBS/CDS ready for insertion promoter (fw-insert) | ||
| D10 | pJ23100 promoter fw-insert | pJ23100 (MP1.1 & MP1.2) | EcoI | SpeI | 58 | J23100 promoter ready for use as a fw-insert | ||
| D11 | pJ23107 ES vector | pJ23107 (MP2.1, MP2.2, MP2.3 & MP2.4) | EcoRI | SpeI | 2096 | Vector with mRFP RBS/CDS ready for insertion of a fw-insert promoter | ||
| D12 | pJ23107 promoter fw-insert | pJ23107 (MP2.1, MP2.2, MP2.3 & MP2.4) | EcoRI | SpeI | 58 | J23107 promoter ready for usage as a fw-insert | ||
| D13 | mRFP | pJ23107 (MP2.2, MP2.3 & MP2.4) | SpeI | PstI | 887 | mRFP RBS/CDS/T sequence, can it be used as a backward insert? => yes but you will not be able to add any biobrick after it | ||
| D14 | Cre ORF | J61047 MP9.1 & MP9.2 | XbaI | PstI | 1063 | Cre ORF extracted for usage as a bw-insert | ||
| D15 | B0030 bw-vector | B0030 MP5.1 & MP5.2 | SpeI | PstI | 2076 | bacward vector for cloning a CDS bw-insert after the B0030 RBS | ||
| D16 | pSB1AK3 vector | B0015 MP3.1 & MP3.2 | EcoRI | PstI | 3148 | pSB1AK3 open vector for BioBrick cloning | ||
| D17 | pSB1A2 E0422 vector | E0422 (MP6.1 & MP6.2) | EcoRI | PstI | 2038 | pSB1A2 open vector for BioBrick cloning | ||
| D18 | BB dig. lox66DapAE.coli | lox66-DapAE.coli P5 PCR product | EcoRI | PstI | ~960 | PCR product digested for cloning as a BioBrick | ||
| D19 | BB dig. lox66DapAsubtilis | lox66-DapAsubtilis P6 PCR product | EcoRI | PstI | ~960 | PCR product digested for cloning as a BioBrick | ||
| D20 | fw insert lox66-DapAE.coli | lox66-DapAE.coli P5 PCR product | XbaI | PstI | ~960 | PCR product digested for usage as a forward insert | ||
| D21 | fw insert lox66-DapAsubtilis | lox66-DapAsubtilis P6 PCR product | XbaI | PstI | ~960 | PCR product digested for usage as a forward insert | ||
July 22
Digestion products
Purification of the digestion products of 21/07/07 purif. using the SV wizard gel cleanup system
elution of the purification products in 30-40microL H2O
Overnight cultures launched
The following ON cultures were launched:
- pJ23107 clone 3 (from the glycerol stock S14.3) in 5ml LB-Amp. For performing a miniprep
- w121 (from glycerol stock S12) in 5 ml LB-Erythromycin. For performing growth-kinetics analysis on DAP supplemented media
I was unable to launch the following culture because of lack of a glycerol stock:
- pJ23107 clone 2 (from the glycerol stock S6.2) in 5ml LB-Amp. For performing a miniprep
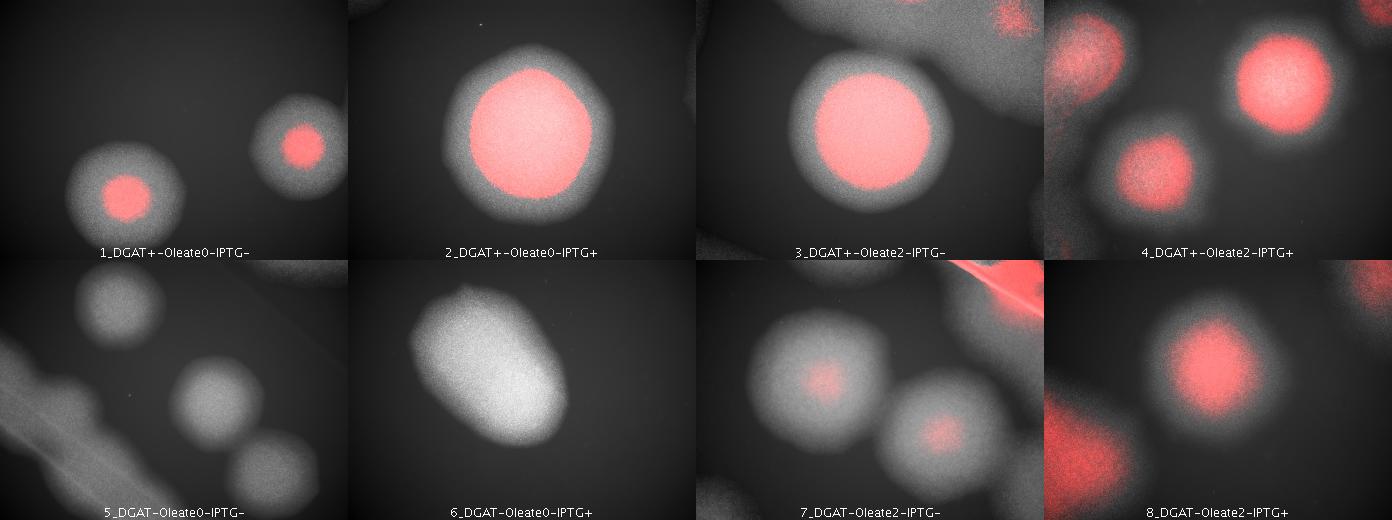
E.Coli pKS::DGAT
We look under microscopy 72 hours after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See July 18).
- Observation:
We can observe E.coli single cells (100X).
- Interpretation:
We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. Signal would be better with more cells.
July 23
w121 growth
w121 culture launched on 22/07/07 did not grow ON: I had forgot to add DAP to LB-Erythro growth medium.
at 12h30, the following culture was launched:
- w121 in 2ml LB-Erythro-DAP
We were unable to perform growth kinetics analysis of w121 in different DAP-supplemented media (because the machine is occupied for the night
Transduction of DapA deletion in MG1655
- The transduction seems to have worked: isolation of clones on LB and LB+DAP for verification.
Migration of digestion products
MiniPrep
Digestion reactions
Each of the digestion reactions that follow were performed as follows:
- 20µL DNA solution (miniprep or PCR product)
- 5µL Buffer 10x
- 0.5µL BSA 100x
- 2µL of each of the 2 enzymes indicated
- H20 qsp 50µL
NEB3 buffer used for XbaI/PstI double digestion NEBEcoRI buffer used for both EcoRI/SpeI & EcoRI/PstI double digestion reactions
| Digestion Products | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number | Product Name | Matrix Name | Enzyme 1 | Enzyme 2 | size | Description | ||
| D22 | pSB1A2 open vector | BBa_J61047 (MP9.1 & MP9.2) | EcoI | PstI | Open pSB1A2 vector for BioBrick EcoRI/PstI insertio | |||
| D23 | AraC/pBad promoter fw-insert | BBa_I0500 (MP4.1 & MP4.2) | EcoI | SpeI | AraC/pBad promoter ready for use as a forward insert | |||
| D24 | eCFP bw-insert | BBa_E0422 (MP6.1 & MP6.2) | XbaI | PstI | eCFP (RBS-OFR_LVA-T) ready for use as a backward insert | |||
| D25 | PoPs to GFP converter | BBa_E0421 (MP7.1 & MP7.2) | XbaI | PstI | PoPs to GFP converter ready for use as a backward insert | |||
| D26 | gfp-tripart | BBa_E0840 (MP8.1 & MP8.2) | XbaI | PstI | gfp-tri part (strongRBS-ORF-T) ready for use as a backward insert | |||
| D27 | lox71-ftsZ bw-insert | lox71-ftsZ PCR product P7 | XbaI | PstI | lox71-ftsZ(unmutated) ready for use as a backward insert | |||
PCRs
| PCR : Lox71-FtsA-FtsZ | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2058 | |||
| 60 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | |||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : Assembly PCR Lox71-FtsA-FtsZ-1 + FtsZ-2 | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 2058 | |||
| 70°C (5x) without oligos + 60°C (35x) | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | 3 Lox71-FtsA-F | 2.5µL | |||
| 3m00' | Oligo R 10µM | 2 FtsZ-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 40x | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | 8.5µl Lox71-FtsA-FtsZ-1 (~50ng) + 17.5µl FtsZ-2 (~50ng) | |||||
| PCR : B0030-DapAColi | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 952 | |||
| 60°C | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | o20 RBS-DapAColi | 2.5µL | |||
| 3m00' | Oligo R 10µM | o7 DapAColi-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35x | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : B0030-DapASubtilis | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 946 | |||
| 60°C | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | o20 RBS-DapASubtilis | 2.5µL | |||
| 3m00' | Oligo R 10µM | o9 DapASubtilis-R | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 35x | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
PCR mutagenesis DGAT
DGAT has Pst1 restriction site. We want to mutagenize it to do a biobrick.
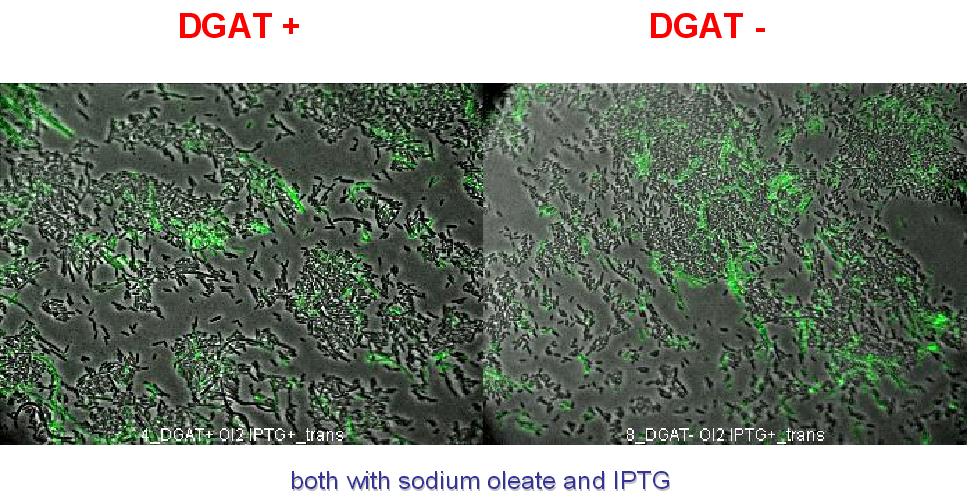
E.Coli pKS::DGAT
We look under microscopy 5 days after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See July 18).
- Observation:
We can observe E.coli single cells (100X).
- Interpretation:
We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. We could think that only dead cells are fluorescent (DGAT could kill the cells) but with cell death marker (green) we can see that cell death is not increased with DGAT.
July 24
Minipreps
BBa_E0422 in pSB1A2 - Clone 1&2 - MP6.1' & MP6.2'
BBa_E0241 in pSB1A2 - Clone 1 - MP7.1'
BBa_J61047 in pSB1A2 - Clone 1&2 - MP9.1' & MP9.2'
Ligation & transformation reactions
| Ligations | |||||
|---|---|---|---|---|---|
| Number | Insert | Insert Volume | Vector | Vector Volume | Comments |
| L1 | D23 (AraC/pBad promoter FI) | D9 (J61002 ready for insertion of a FI) | |||
| L2 | D18 (BB dig. lox66DapAE.coli) | D22 (pSB1A2 Eco, Pst) | |||
| L3 | D19 (BB dig. lox66DapAsubtilis) | D22 (pSB1A2 Eco, Pst) | |||
| L4 | D20 (Lox66-DapAE.coli BI) | D1 (pJ23100 BV) | |||
| L5 | D21 (lox66-DapAsubtilis BI) | D1 (pJ23100 BV) | |||
| L6 | D20 (Lox66-DapAE.coli BI) | D3 (pJ23107 BV) | |||
| L7 | D21 (lox66-DapAsubtilis BI) | D3 (pJ23107 BV) | |||
| L8 | D14 (Cre ORF) | D15 (B0030 BV) | |||
| L9 | D27 (Lox71-ftsZ BI) | D1 (pJ23100 BV) | |||
| L10 | D27 (Lox71-ftsZ BI) | D3 (pJ23107 BV) | |||
- lox66 (annealing O14 & O15) & lox71 (annealing O16 & O17) in D22 (pSB1A2 Eco+Pst)
Annealing of Lox66 and Lox71
Lox66 and Lox71 oligos were designed to form a double-stranded DNA. The extremities bear cohesive overhangs corresponding to digestion by EcoRI and SpeI. See oligos 14 + 15 and 16 + 17 here.
Annealing mix:
- 8 μL of each of the concentrated primers
- 4 μL of salt solution (10 mM NaCl)
- 20 μL of water
Mix in a PCR tube to a beaker of boiling water and just allow the water to cool down naturally.
More information [http://openwetware.org/wiki/Annealing_complementary_primers here]
Purification of PCR Products
- RBS-DapAColi = P8
- RBS-DapASubtilis = P9
- Lox71-FtsA-FtsZ = P10
- DGAT1 = P3
- DGAT2 = P4
July 25
Digestions
Digestion of purifications of PCR products
| Digestion Products | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number | Product Name | Matrix Name | Enzyme 1 | Enzyme 2 | size | Description | ||
| D28 | RBS-DapAColi BB | P8 (RBS-DapAColi) | EcoRI | PstI | ~950 | Biobrick | ||
| D29 | RBS-DapAColi | P8 (RBS-DapAColi) | XbaI | PstI | ~950 | Backward insert | ||
| D30 | RBS-DapASubtilis BB | P9 (RBS-DapASubtilis) | EcoRI | PstI | ~950 | Biobrick | ||
| D31 | RBS-DapASubtilis | P9 (RBS-DapASubtilis) | XbaI | PstI | ~950 | Backward insert | ||
| D32 | Lox71-FtsA-FtsZ | P10 Lox71-FtsA-FtsZ | XbaI | PstI | ~2600 | Backward insert - EcoRI site still presents : we can't use EcoRI | ||
Ligation & transformation reactions
The Ligation reactions performed yesterday have all failed (the reactions were performed at 42° by mistake, then, another 1µL of ligase was added for a 5min rxn at 25°)
The following reactions were performed today
| Ligations | |||||
|---|---|---|---|---|---|
| Number | Insert | Insert Volume (µL) | Vector | Vector Volume (µL) | Comments |
| L1' | D23.1 (AraC/pBad promoter FI) | 8.5 | D9.1 (J61002 ready for insertion of a FI) | 1.5 | construct with pBad promotor regulating expression of mRFP (in order to test activity of pBad) |
| L2' | D18 (BB dig. lox66DapAE.coli) | 7 | D22.1 (pSB1A2 Eco, Pst) | 3 | Insertion of lox66DapAE.coli construct as a biobrick in pSB1A2 plasmid |
| L3' | D19 (BB dig. lox66DapAsubtilis) | 7 | D22.1 (pSB1A2 Eco, Pst) | 3 | Insertion of lox66DapAsubtilis construct as a biobrick in pSB1A2 plasmid |
| L4' | D20 (Lox66-DapAE.coli BI) | 8 | D1 (pJ23100 BV) | 2 | lo66-DapAE.coli under regulation of strong promotor pJ23100 |
| L5' | D21 (lox66-DapAsubtilis BI) | 6.5 | D1 (pJ23100 BV) | 2 (+1.5µL H2O) | lox66-DapAsubtilis under regulation of strong promotor pJ23100 |
| L6' | D14.1 (Cre ORF) | 8 | D15.1 (B0030 BV) | 2 | construction of the biobrick strong RBS(B0030)-Cre ORF |
| L7' | D27 (Lox71-ftsZ BI) | 6 | D1 (pJ23100 BV) | 2 (+2µL H2O) | lox71-ftsZ under regulation of strong promotor pJ23100 |
| L8' | D27 (Lox71-ftsZ BI) | 6 | D3 (pJ23107 BV) | 2 (+2 H2O) | lox71-ftsZ under regulation of promotor pJ23107 (intermediate strength) |
| L9' | lox66 [O14+O15] (0.2µM) | 2 | D22.1 (pSB1A2 Eco, Pst) | 2 (+6 H2O) | lox66 cloned as a biobrick in pSB1A2 |
| L10' | lox71 [O16+O17] (0.2µM) | 2 | D22.1 (pSB1A2 Eco, Pst) | 2 (+6 H2O) | lox71 cloned as a biobrick |
July 26
Colonies PCR for cloning verification
L1.1, L1.2, L2.1, L2.2, L9.1, L10.1
L6: 8 clones
Toothpick in 50µl sterile water ==> 10'@100°C
PCR Mix:
- 212.5 µl Master Mix
- 8.5 µl VF2
- 8.5 µl VR
12 µl of the mix in 13µl of lysed cells for each PCR reaction
Cycle x30:
- 95°C 30
- 60°C 30
- 68°C 2'
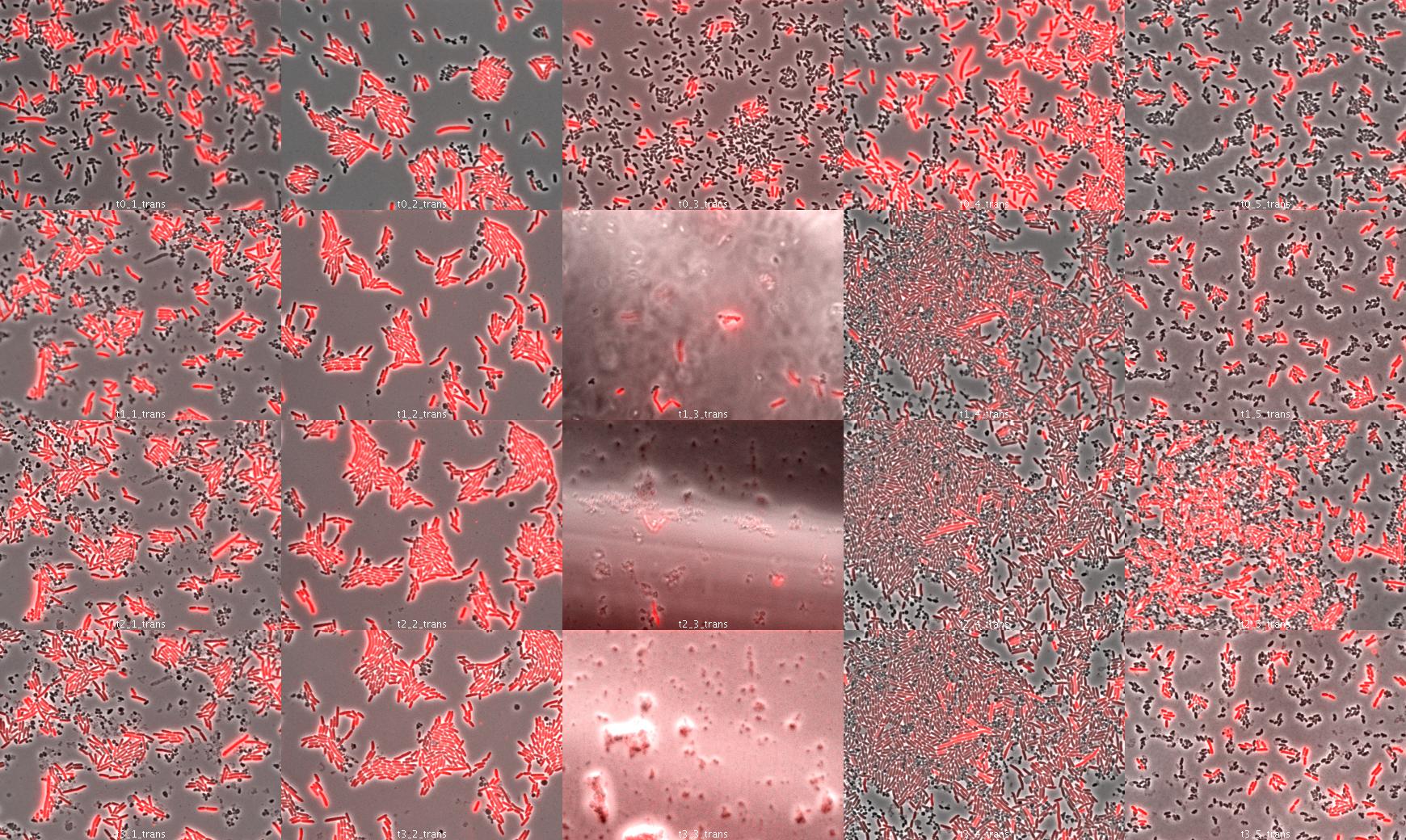
Microscopic DAP complementation experiment
We used wild type E.coli transformed by part J23107 (RFP) and dapA- E.coli (w121) auxotroph to DAP. E.coli transformed by part J23107 (RFP) is able to synthesize DAP and export it. We suppose dap- E.coli could be feed by wt E.coli.
We tried to observe kinetics by fluorescence microscopy. We let separately in culture overnight dapA- E.coli (LB/erythromycine/DAP) and rfp wt E.coli (LB ampicillin). The day after, we diluted both liquid culture 1/200 to have exponential culture.
Then we put on slide different conditions:
- 50% wt rfp / 50% dapA- (colomn 1)
- 70% wt rfp / 30% dapA- (colomn 2)
- 30% wt rfp / 70% dapA- (colomn 3)
- 90% wt rfp / 10% dapA- (colomn 4)
- 10% wt rfp / 90% dapA- (colomn 5)
Under microscope and at 37°C we observed every 30 minutes the bacteria:
The dapA- bacteria do not look to survive; we can observe natural selection directly: rfp wt cells have higher fitness: they multiply
much more than dapA- cells. If dapA- cells were growing, it would have been as parasites because wt cells do not need dapA- cells. So this is just a preliminary experiment because in our system, cells able to multiply need cells unable because of high dapA secretion. This was to see how many wt cells are needed to dapA cells to survive.
We were observing cells on slide under microscope. They do not look to multiply very fast (3-4 divisions in 2 hours). It is possible that dapA cells do not survive because they already are stressed by culture conditions. So we would like to do the same in liquid conditions (we take samples every hour).
July 27
PCR to perform
The following PCR reactions have been performed
- P5
- P6
- P8
- P9
Oligo design for Wanner deletion
We were for now on not able to perform the deletion of DapA through transduction. So we will try the deletion through the Wanner method. We will also attempt the deletion of FtsZ.
We will use the protocol of the keio collection: [http://ecoli.naist.jp/gb6/Resources/deletion/Del_construct.html]
- 20nt of upstream region of kan gene,
5'-ATTCCGGGGATCCGTCGACC-3' (P1)
- 20nt of kan downstream sequence,
5'-TGTAGGCTGGAGCTGCTTCG-3' (P2)
DapA deletion
- 50nt DapA upstream region
5'-CATACCAAACGTACCATTGAGACACTTGTTTGCACAGAGGATGGCCCATG-3' (P3)
- 50nt DapA downstream region (reverse complement)
5'-AAGCCATCAAATCTCCCTAAACTTTACAGCAAACCGGCATGCTTAAGCGC-3' (P4)
- dDapA-F
CATACCAAACGTACCATTGAGACACTTGTTTGCACAGAGGATGGCCCATGATTCCGGGGATCCGTCGACC (P3 + P1)
- dDapA-R
AAGCCATCAAATCTCCCTAAACTTTACAGCAAACCGGCATGCTTAAGCGCTGTAGGCTGGAGCTGCTTCG (P6 + P2)
FtsZ deletion
- 50nt FtsZ upstream region
5'-GACGATGATTACGGCCTCAGGCGACAGGCACAAATCGGAGAGAAACTATG-3' (P5)
- 50nt FtsZ downstream region (reverse complement)
5'-GAAACCCAAATTCCAGTCAATTCTTAATCAGCTTGCTTACGCAGGAATGC-3' (P6)
- dFtsZ-F
GACGATGATTACGGCCTCAGGCGACAGGCACAAATCGGAGAGAAACTATGATTCCGGGGATCCGTCGACC (P5 + P1)
- dFtsZ-R
GAAACCCAAATTCCAGTCAATTCTTAATCAGCTTGCTTACGCAGGAATGCTGTAGGCTGGAGCTGCTTCG (P6 + P2)
Ligations
| Ligations | ||||||
|---|---|---|---|---|---|---|
| Number | Insert | Insert Volume (µL) | Vector | Vector Volume (µL) | Comments | Number of colonies |
| Control 0 | pUC19 | control of transformation efficiency | 4 | |||
| Control 1 | MP3.1 digested with EcoRI (D0) | 2 | control of ligation efficiency | ~80 | ||
| Control 2 | MP3.1 digested with EcoRI (D0) | 2 | without ligase in the mix: control of digestion efficiency | 0 | ||
| Control 3 | D9.1 (J61002 ready for insertion of a FI) | 2µl | control of D9.2 digestion | 6 | ||
| Control 4 | D15.1 (B0030 BV) | 2µl | control of D15.2 digestion | 1 | ||
| Control 5 | D22.2 (pSB1A2 Eco, Pst) | 2µl | control of D22.1 digestion | 0 | ||
| L1 | D23.2 (AraC/pBad promoter FI) | 8 | D9.1 (J61002 ready for insertion of a FI) | 2 | construct with pBad promotor regulating expression of mRFP (in order to test activity of pBad) | 12 |
| L6 | D14.2(Cre ORF) | 8 | D15.1 (B0030 BV) | 2 | construction of the biobrick strong RBS(B0030)-Cre ORF | ~150 |
| L9 | lox66 [O14+O15] (0.2µM) | 2 | D22.2 (pSB1A2 Eco, Pst) | 2 (+6 H2O) | lox66 cloned as a biobrick in pSB1A2 | 0 |
| L10 | lox71 [O16+O17] (0.2µM) | 2 | D22.2 (pSB1A2 Eco, Pst) | 2 (+6 H2O) | lox71 cloned as a biobrick | 0 |
Ligation mix:
- 2µl TP
- 1µl Ligase
- 7µl H20
Reaction in 20µl
ON @ 4°C
Minipreps
minipreps were performed on the following ON cultures:
cultures of different clones of the ligation reactions of 25/07/07
- L1'.2 & L1'.3
- L2'.1 & L2'.2
- L9'
- L10'
- L6'.1, L6'.2 & L6'.3
for more detail on these ligation reactions, go to 25/07/07
no glycerol stocks of hese strains have been generated yet (the culyures are launched tonight in order to do this)
minipreps & glycerol stocks:
- MP12.1 & MP12.2 (2 clones after transformation with biobrick BBa_P1003)
July 28
Transformations
Transformations of 5µl from yesterday's ligations in DH5alpha subcloning efficiency competant cells (invitrogen).
Control: 2.5µl of pUC19 (100pg/µl)
July 29
July 30
Sequences
All the biobricks that we got from the plate and all the ligations that succeeded up to now.
Colonies PCR screen
On colony PCRs performed to screen for correct ligation reactions (performed on 27/07/07):
10 clones of L1 ligation reactions, 6 clones of L6 ligation reactions
preparation of the following PCR mix
- 18x (12.5µL 2xreaction mix)= 225
- 18x (0.5µL oligo O18 VF2 10µM)=9
- 18x (0.5µL oligo O19 VR 10µM)=9
- 12µL H20+colony (10min 100°)
PCRs
| PCR : P5 lox66-DapAColi | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 985 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | O6 | 2.5µL | |||
| 2m00' | Oligo R 10µM | O7 | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 30 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : P6 Lox66-DapASubtilis | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 979 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | O8 | 2.5µL | |||
| 2m00' | Oligo R 10µM | O9 | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 30 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | 0.5µl Subtilis DNA | |||||
| PCR : P7 Lox71-FtsZ | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 1260 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | O1 | 2.5µL | |||
| 2m00' | Oligo R 10µM | O2 | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 30 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : P8 RBS-DapAColi | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 952 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | O20 | 2.5µL | |||
| 2m00' | Oligo R 10µM | O7 | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 30 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | Toothpick in MG16655 Glycerol | |||||
| PCR : P11 Lox71-KmR-Lox66 | ||||||
|---|---|---|---|---|---|---|
| PCR Settings | Buffer (5x) | 5x 10µL | Expected size | |||
| Annealing (°C) | MgCl2 10µM | 10µM 0µL | 1084 | |||
| 55 | dNTP 10µM | 10µM 1µL | Success | |||
| Time Elongation | Oligo F 10µM | O10 | 2.5µL | |||
| 2m00' | Oligo R 10µM | O11 | 2.5µL | Image (click to enlarge) | ||
| Number cycles | Water | 34µl | [[Image:|30px]] | |||
| 30 | Polymerase | Phusion 0.5µL | Band (0=ladder) | |||
| DNA | 0.5 µl MP12 | |||||
Digestion reactions
The following digestion reactions have been performed:
| Digestion Products | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number | Product Name | Matrix Name | Enzyme 1 | Enzyme 2 | size | Description | ||
| D19' | BB dig. lox66DapAsubtilis | lox66-DapAsubtilis P6' PCR product | EcoRI | PstI | ~960 | PCR product digested for cloning as a BioBrick | ||
| D21' | bw-insert lox66-DapAsubtilis | lox66-DapAsubtilis P6' PCR product | XbaI | PstI | ~960 | PCR product digested for cloning as a BI | ||
| D27' | lox71-ftsZ bw-insert | lox71-ftsZ PCR product P7 | XbaI | PstI | lox71-ftsZ(unmutated) ready for use as a backward insert | |||
| D33 | RBS(B0030)CreORF | L6' (clone 1 & 2) | XbaI | PstI | RBS-Cre digestion for use as a BI | |||
| D34 | lox66 | L9' lox66 | SpeI | PstI | Biobrick plasmid carrying lox66 digested as a BV | |||
| D35 | lox71 | L10' lox71 | SpeI | PstI | Biobrick plasmid carrying lox71 digested as a BV | |||
Ligations
| Ligations | ||||||
|---|---|---|---|---|---|---|
| Number | Insert | Insert Volume (µL) | Vector | Vector Volume (µL) | Comments | Number of colonies |
| Control 1 | MP3.1 digested with EcoRI (D0) | 2 | control of ligation efficiency | |||
| a faire | MP3.1 digested with EcoRI (D0) | 2 | without ligase in the mix: control of digestion efficiency | |||
| Control 2 | D35 | 2µl | control of D35 digestion | 1 | ||
| Control 3 | D34 | 2µl | control of D34 digestion | 2 | ||
| L3 | D19 (lox66dapAsubtilis) | 4 | D22.1 (pSB1A2 dig. EcoRI&PstI) | 2 | Insertion of lox66DapASubtilis as a biobrick | |
| L5 | D21’(lox66DapASubtilis) | 4 | D1 (j23100 digested for CDS insertion) | 2 | construction of the biobrick J23100-lox66dapAsubtilis | 0 |
| L7 | D27' (lox-ftsZ) | 4 | D1 (j23100 digested for CDS insertion) | 2 | construction of the biobrick J23100- lox-ftsZ | 0 |
| L8 | D27' (lox-ftsZ) | 4 | D3 (j23107 digested for CDS insertion) | 2 | construction of the biobrick J23107- lox-ftsZ | 0 |
| L9 | lox66 [O14+O15] (2µM) | 2 | D22.2 (pSB1A2 Eco, Pst) | 2 | lox66 cloned as a biobrick in pSB1A2 | |
| L10 | lox71 [O16+O17] (2µM) | 2 | D22.2 (pSB1A2 dig. EcoRI & PstI) | 2 | lox71 cloned as a biobrick in pSB1A2 | 0 |
| L11 | D21’(lox66dapAsubtilis BI) | 4 | D3 (j23107 digested for CDS insertion) | 2 | construction of the biobrick: J23107 promotor-lox66dapAsubtilis | 1 |
| L12 | D26.1 (gfp-tripart BI) | 4 | D35 (lox71 plasmid digested as a BV) | 2 | construction of the biobrick lox71-gfp | 14 |
| L13 | D13.2 (mRFP de pJ23100) | 4 | D34 (lox66 plasmid digested as a BV) | 2 | construction of the biobrick lox66-mRFP | 0 |
| L14 | D33.1 (RBS-Cre BI) | 4 | D1 (j23100 digested for CDS insertion) | 2 | J23100-RBS-Cre | |
| L15 | D33.1 (RBS-Cre BI) | 4 | D3 (j23107 digested for CDS insertion) | 2 | J23107-RBS-Cre | 0 |
| L16 | D32 (lox71-ftsAftsZ BI) | 4 | D1 (j23100 digested for CDS insertion) | 2 | J23100-lox71-ftsAftsZ | 0 |
| L17 | D32 (lox71-ftsAftsZ BI) | 4 | D3 (j23107 digested for CDS insertion) | 2 | J23107-lox71-ftsAftsZ | 0 |
| L18 | D31 (RBSdapAsubtilis BI) | 4 | D1 (j23100 digested for CDS insertion) | 2 | J23100-RBSdapAsubtilis | 0 |
| L19 | D31 (RBSdapAsubtilis BI) | 4 | D3 (j23107 digested for CDS insertion) | 2 | J23107-RBSdapAsubtilis | 0 |
| L20 | D30 (RBSdapAsubtilis dig. BB) | 4 | D22.2 (pSB1A2 Eco, Pst) | 2 | 0 | |
| L21 | D29 | 4 | D1 (j23100 digested for CDS insertion) | 2 | 0 | |
| L22 | D29 | 4 | D3 (j23107 digested for CDS insertion) | 2 | 0 | |
| L23 | D28 (RBSdapAcoli dig.BB) | 4 | D22.2 (pSB1A2 Eco, Pst) | 2 | 1 | |
| L24 | D21’(lox66dapAsubtilis BI) | 4 | D3 (j23107 digested for CDS insertion) | 2 | 0 | |
| L25 | D36 (lox71-KmR-lox66 dig. BB) | 4 | D22.2 (pSB1A2 dig. EcoRI & PstI) | 2 | 1 | |
Ligation mix:
- 1µl 10x buffer
- 0.5µl Ligase
- 2µL plasmid DNA solution
- 4µL insert DNA solution
Reaction in 10µl
ON @ 4°C
MiniPreps
- L1".1, L1".2
- L6".1, L6".2
Sequencing
The following sequencing reactions have been performed
DB_MP1.1+O18 DB_MP1.1+O19 DB_MP1.2+O18 DB_MP1.2+O19 DB_MP2.1+O18 DB_MP2.1+O19 DB_MP3.1+O18 DB_MP3.1+O19 DB_MP3.2+O18 DB_MP3.2+O19 DB_MP4.1+O18 DB_MP4.1+O19 DB_MP4.2+O18 DB_MP4.2+O19 DB_MP5.1+O18 DB_MP5.1+O19 DB_MP5.2+O18 DB_MP5.2+O19 DB_MP6.1+O18 DB_MP6.1+O19 DB_MP6.2+O18 DB_MP6.2+O19 DB_MP7.1+O18 DB_MP7.1+O19 DB_MP7.2+O18 DB_MP7.2+O19 DB_MP8.1+O18 DB_MP8.1+O19 DB_MP8.2+O18 DB_MP8.2+O19 DB_MP9.1+O18 DB_MP9.1+O19 DB_MP9.2+O18 DB_MP9.2+O19 DB_MP10.1+O18 DB_MP10.1+O19 DB_MP10.2+O18 DB_MP10.2+O19 DB_MP11.3+O18 DB_MP11.3+O19 DB_MP11.4+O18 DB_MP11.4+O19 DB_MP12.1+O18 DB_MP12.1+O19 DB_MP12.2+O18 DB_MP12.2+O19 DB_L1.2+O18 DB_L1.2+O19 DB_L1.3+O18 DB_L1.3+O19 DB_L2.1+O18 DB_L2.1+O19 DB_L2.2+O18 DB_L2.2+O19 DB_L9+O18 DB_L9+O19 DB_L10+O18 DB_L10+O19 DB_L6.1+O18 DB_L6.1+O19 DB_L6.2+O18 DB_L6.2+O19 DB_L6.3+O18 DB_L6.3+O19
July 31
pTOPO cloning
Cloning of PCR products: P5, P6, P7, P8, P9, P10 (see Paris/Freezer)
Transformation
Ligations from yesterday
PCR mutagenesis DGAT
DGAT has Pst1 restriction site. We want to mutagenize it to do a biobrick.